
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,780,667 – 22,780,784 |
| Length | 117 |
| Max. P | 0.547973 |

| Location | 22,780,667 – 22,780,766 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -26.03 |
| Energy contribution | -28.02 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
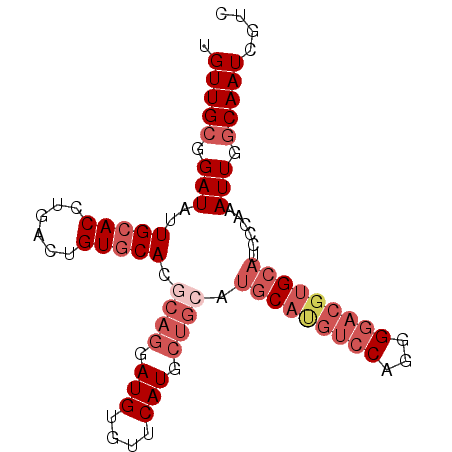
>3L_DroMel_CAF1 22780667 99 - 23771897 UGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGCCUGUGUGCGGUUGAAUUGCUGACAACAC .(((((..(((((((.(((....))).))))((((((((((...))))))))))........(((....))).)))..)))))................ ( -40.40) >DroPse_CAF1 51607 93 - 1 UGACUGUACACGCAGGAUGUGUCCAUACUGAAUACUGCAU------CCGUGCAUCCCCAAUUGGCAAUCGUCUGUGUGCGGUUGAAUUGCUGACAACAC .(((((((((((((((((((................))))------)).)))..........(((....))).))))))))))................ ( -26.49) >DroSec_CAF1 43455 99 - 1 UGACUGUGCACGCAGGAUGUGUUCAUGCUGCCUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUCUGUGUGCGGUUGAAUUGCUGACAACAC .(((((..(((((((.(((....))).)))).(((((((((...))))))))).........(((....))).)))..)))))................ ( -36.00) >DroSim_CAF1 45542 99 - 1 UGACUGUGCACGCAGGAUGCGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUCUGUGUGCGGUUGAAUUGCUGACAACAC .(((((..(((((((.(((....))).))))((((((((((...))))))))))........(((....))).)))..)))))................ ( -36.70) >DroEre_CAF1 42633 99 - 1 UGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUCUGUGUGCGGUUGAAUUGCUGACAACAC .(((((..(((((((.(((....))).))))((((((((((...))))))))))........(((....))).)))..)))))................ ( -37.70) >DroYak_CAF1 46002 84 - 1 --------CACGCAGGAUGUGUUCAU-------GCACGUCCAGGGGACGUGCAUCCAAAAUUGGCAAUCGUCUAUGUGUGGUUGAAUUGCCAACAACAC --------(((((((((((.....((-------((((((((...))))))))))(((....)))....))))).))))))((((......))))..... ( -30.80) >consensus UGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUCUGUGUGCGGUUGAAUUGCUGACAACAC ......((((((((((...((((.((.....((((((((((...)))))))))).....)).))))....))))))))))((((.........)))).. (-26.03 = -28.02 + 1.99)



| Location | 22,780,694 – 22,780,784 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -21.28 |
| Energy contribution | -23.67 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22780694 90 - 23771897 UGUUGCGGAUAUUGCACCUGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGCC .(((((.(((..(((((......))))).((((.(((....))).))))((((((((((...)))))))))).....))).))))).... ( -35.20) >DroPse_CAF1 51634 84 - 1 UGUUGCGGAUAUUGCACCUGACUGUACACGCAGGAUGUGUCCAUACUGAAUACUGCAU------CCGUGCAUCCCCAAUUGGCAAUCGUC .(((((.(((..(((((.....(((....)))((((((................))))------)))))))......))).))))).... ( -18.99) >DroSec_CAF1 43482 90 - 1 UGUUGCGGAUAUUGCACCUGACUGUGCACGCAGGAUGUGUUCAUGCUGCCUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUC .(((((.(((..(((((......))))).((((.(((....))).)))).(((((((((...)))))))))......))).))))).... ( -33.50) >DroSim_CAF1 45569 90 - 1 UGUUGAGGAUAUUGCACCUGACUGUGCACGCAGGAUGCGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUC (((..(((((..(((((......))))).((((.(((....))).))))..((((((((...))))))))))))....)..)))...... ( -32.30) >DroEre_CAF1 42660 90 - 1 UGUUGCGGAUAUUGCACCUGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUC .(((((.(((..(((((......))))).((((.(((....))).))))((((((((((...)))))))))).....))).))))).... ( -35.20) >DroYak_CAF1 46029 71 - 1 UGUUGCUGAUAUUG------------CACGCAGGAUGUGUUCAU-------GCACGUCCAGGGGACGUGCAUCCAAAAUUGGCAAUCGUC .(((((..((..((------------(((...((((((((....-------)))))))).(....))))))......))..))))).... ( -28.50) >consensus UGUUGCGGAUAUUGCACCUGACUGUGCACGCAGGAUGUGUUCAUGCUGCAUGCAUGUCCAGGGGACGUGCAUCCCAAAUUGGCAAUCGUC .(((((.(((..(((((......))))).((((.(((....))).)))).(((((((((...)))))))))......))).))))).... (-21.28 = -23.67 + 2.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:18 2006