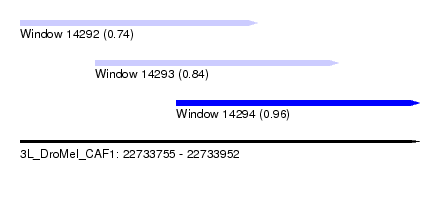
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,733,755 – 22,733,952 |
| Length | 197 |
| Max. P | 0.955151 |

| Location | 22,733,755 – 22,733,872 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -41.43 |
| Energy contribution | -41.65 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22733755 117 + 23771897 GUA---AGUUAUCUGUAUCUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCUUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG ...---....(((.((((.((((((((((....))))))).))).)))).)))((((((.(.(((((..((....))(((((((.(....).)))))))))))).)))))))........ ( -46.60) >DroVir_CAF1 13740 114 + 1 UGG---AUGUGU---UAACUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG .((---((((((---.....(.(((((((....))))))))))))))......((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).))..... ( -48.70) >DroGri_CAF1 13731 114 + 1 UGG---AUGUGU---UAACUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG .((---((((((---.....(.(((((((....))))))))))))))......((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).))..... ( -48.70) >DroWil_CAF1 17790 110 + 1 UUA---AGUUGUC-------GUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG ...---(((((((-------(.(((((((....))))))))).))).)))...((((((.(.(((((..((....))(((((((.(....).)))))))))))).)))))))........ ( -48.00) >DroMoj_CAF1 14517 114 + 1 UGG---AUGUGU---UAACUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG .((---((((((---.....(.(((((((....))))))))))))))......((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).))..... ( -48.70) >DroAna_CAF1 11737 119 + 1 GGAAGAAGUUAUCUAAAUCUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAAGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAAGGCUUCUCACCC-AAUG ((.((((((.........((.((((..(((((((((((((.((((.((((....((((.....)))).))))...)))).)))).))).))))))..)))).))))))))...))-.... ( -43.60) >consensus UGA___AGGUGU___UAACUGUCGCUCGCCUUGGUGGGUGCGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUG ....................(.(((((((....))))))))((....))....((((((.(.(((((..((....))(((((((.(....).)))))))))))).)))))))........ (-41.43 = -41.65 + 0.22)


| Location | 22,733,792 – 22,733,912 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -43.99 |
| Energy contribution | -44.68 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22733792 120 + 23771897 CGCACAUGCUGAUUGAGGAUGGCUUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGGUUGAGAAUGGCGCGCUGCCGAAAA .((((((((....((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).((....))....))))))))..........((((.....)))).... ( -47.00) >DroGri_CAF1 13765 120 + 1 CGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGUAUCGCGCGCUGCCGGAAA .((((((((....((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).((....))....))))))))..........(((.((...)))))... ( -47.20) >DroSim_CAF1 11674 120 + 1 CGCACAUGCUGAUUGAGGAUGGCUUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGAAUGGCGCGCUGCCGAAAG .((((((((....((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).((....))....))))))))..........((((.....)))).... ( -47.00) >DroWil_CAF1 17820 120 + 1 CGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUAUGCUGAUUGAGUAUGGCUCGCUGUCGAAAG ....(((..((..((((((.(.(((((..((....))(((((((.(....).)))))))))))).)))))))..)).)))((((((..((((((.....))))))...))).)))..... ( -45.60) >DroMoj_CAF1 14551 120 + 1 CGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGUAUGGCUCGCUGCCGGAAG .((((((((....((((((.(.(((((..((....))(((((((.(....).)))))))))))).))))))).((....))....))))))))..........((((.....)))).... ( -50.60) >DroAna_CAF1 11777 118 + 1 CGCACAUGCUGAUUGAAGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAAGGCUUCUCACCC-AAUGGGAGCGUA-AUGCUGGUUAAGAAUGGCGCGCUGCCGGAAG .((.(((.((......)))))))((((((((.....((((((((.(....).))))))))...(((.(((((...-..)))))((((.-...((.....))....))))))))))))))) ( -44.60) >consensus CGCACAUGCUGAUUGAGGAUGGCCUUCUGGCGAAAGUGCUUGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGAAUGGCGCGCUGCCGAAAG .((((((((....((((((.(.(((((..((....))(((((((.(....).)))))))))))).)))))))((.....))....))))))))..........((((.....)))).... (-43.99 = -44.68 + 0.69)


| Location | 22,733,832 – 22,733,952 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -50.91 |
| Consensus MFE | -45.97 |
| Energy contribution | -46.42 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22733832 120 + 23771897 UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGGUUGAGAAUGGCGCGCUGCCGAAAACUGCGCGAGCAGUAUGGGCAGGUGAAGGGCUUUUCGUUGG ((((.(....).))))(((((((((((..(((((...............((((((........))))))..((((.(..(((((....))))).).)))))))))..))))))))))).. ( -52.50) >DroVir_CAF1 13814 120 + 1 UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGGUUGAGUAUGGCGCGCUGCCGGAAGCUGCGCGAGCAGUACGGGCAGGUGAAUGGCUUUUCGUUUG .(((.(....).)))((((((((((((..(((((.........((((.((((((.....))))))..))))((((.(..(((((....))))).).)))))))))..)))))))))))). ( -54.90) >DroGri_CAF1 13805 120 + 1 UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGUAUCGCGCGCUGCCGGAAACUGCGCGAGCAGUACGGACACGUGAAUGGCUUUUCGUUAG ((((.(....).))))(((((((((((..(((((((..(((..(((..((((((.....)))).))..)))..)))...(((((....))))).)))....))))..))))))))))).. ( -49.50) >DroWil_CAF1 17860 120 + 1 UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUAUGCUGAUUGAGUAUGGCUCGCUGUCGAAAGCUGCGCGAACAGUACGGGCAGGUGAAGGGCUUUUCGUUGG ((((.(....).))))(((((((((((..(((((((..((..((((((((((((.....))))))....(((......)))))))))..))...))))....)))..))))))))))).. ( -47.00) >DroMoj_CAF1 14591 120 + 1 UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGUAUGGCUCGCUGCCGGAAGCUGCGCGAGCAGUACGGGCAGGUGAAUGGCUUUUCGUUGG ((((.(....).))))(((((((((((..(((((.........(((..((((((.....))))))...)))((((.(..(((((....))))).).)))))))))..))))))))))).. ( -53.40) >DroAna_CAF1 11817 118 + 1 UGCCGCACUCGGGGCAGGCGAAAGGCUUCUCACCC-AAUGGGAGCGUA-AUGCUGGUUAAGAAUGGCGCGCUGCCGGAAGCUGCGCGAACAGUACGGGCAGGUGAAGGGUUUUUCGUUGG ((((.(....).))))(((((((((((..(((((.-.(((....))).-.((((.((.........(((((.(.(....)).)))))......)).)))))))))..))))))))))).. ( -48.16) >consensus UGCCGCACUCGGGGCAGGCGAAGGGCUUCUCACCCGAAUGGAUGCGUAUGUGCUGAUUGAGUAUGGCGCGCUGCCGGAAGCUGCGCGAGCAGUACGGGCAGGUGAAGGGCUUUUCGUUGG ((((.(....).))))(((((((((((..(((((.........(((..((((((.....))))))...)))((((.(..(((((....))))).).)))))))))..))))))))))).. (-45.97 = -46.42 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:12 2006