
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,623,349 – 2,623,451 |
| Length | 102 |
| Max. P | 0.654990 |

| Location | 2,623,349 – 2,623,451 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.77 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.25 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2623349 102 - 23771897 GUUUAAAUAAUUU----AAU-----GUAGAUUAUAGAGCGUAUAA--AUG-UAGUUUACCUGGACAAGCUGUCCGGCUGCCAGCUGAUAGCUGUUGGGUAACGAGAGUCCCGCA ......(((((((----(..-----.))))))))...(((.....--...-.....((((..(...(((((((.(((.....)))))))))).)..))))((....))..))). ( -27.10) >DroGri_CAF1 13558 97 - 1 ACU---UUUAGCU----GAAUUUGUAUAAU--------A--ACAAAAUCCUCCUCUUACCUGGACAAGCUGUCCUGCGGCGAGCUGAUAGCUGUUGGGCAGCGAGAGAGCCGCA ...---....(((----((.(((((.....--------.--))))).))(((.(((.....)))...(((((((.(((((.........))))).))))))))))..))).... ( -28.30) >DroSec_CAF1 21442 84 - 1 GUUUAAAUAAUUA----AAA-----------------------AA--ACG-UAGUUUACCUGAACAAGCUGUCCGGCUGCCAGCUGAUAGCUAUUGGGCAACGAGAGUCCCGCA (((((...(((((----...-----------------------..--...-)))))....))))).(((((((.(((.....))))))))))...(((..((....)))))... ( -20.50) >DroSim_CAF1 23550 100 - 1 GCUUAAAUAAUUG----AAA-------AAAUGAUAGACCGCAAAA--ACG-UAGUUUACCUGAACAAGCUGUCCGGCUGCCAGCUGAUAGCUAUUGGGCAACGAGAGACCCGCA ((((((.......----...-------......(((((..(....--..)-..)))))........(((((((.(((.....)))))))))).))))))..(....)....... ( -21.60) >DroEre_CAF1 22284 91 - 1 AUUUAAAUAAUUGUU--------------------GAGCGUAAAA--ACG-UAGCUUACCUGAACAAGCUGUCCGGCGGCCAGCUGAUAGCUGUUGGGCAGCGAGAGUCCGGCA ..........(((((--------------------.((.((((..--...-....)))))).)))))(((((((((((((.........)))))))))))))............ ( -29.90) >DroYak_CAF1 25685 107 - 1 U-UAAAUUAAGUGUUUUAAAC---UCCAAUUUUUAGAGCGUAAAA--ACG-UAGCUUACCUGGACAAGCUGUCCGGCAGCCAGCUGAUAGCUGUUGGGCAACGAGAGUCCUGCA .-.....(((((((((((((.---.......))))))))((....--)).-..)))))...((((..(.(((((((((((.........))))))))))).)....)))).... ( -28.50) >consensus GUUUAAAUAAUUG____AAA_______AA______GAGCGUAAAA__ACG_UAGCUUACCUGAACAAGCUGUCCGGCUGCCAGCUGAUAGCUGUUGGGCAACGAGAGUCCCGCA ..........................................................((..(...(((((((.(((.....)))))))))).)..))................ (-16.33 = -16.25 + -0.08)
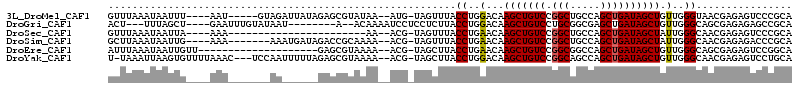
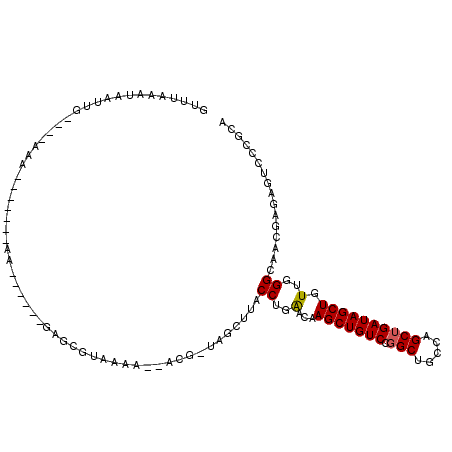
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:18 2006