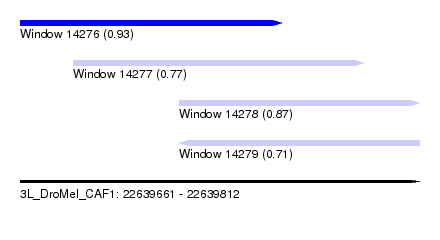
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,639,661 – 22,639,812 |
| Length | 151 |
| Max. P | 0.934341 |

| Location | 22,639,661 – 22,639,760 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22639661 99 + 23771897 G------ACAAACACCGAGCUG--------------GUAUUUUAAAUUCAAUGAUGCGGCAGGCGGUAAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCGAAAG-GACGCCAACU (------(((........((((--------------.((((...........))))))))(((((.....))))).....)))).......(((((((((((.....))-)).))))))) ( -31.30) >DroSec_CAF1 8456 99 + 1 G------ACACACACCGAGCUG--------------GUAUUUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCGAAAG-GACACCAGCC .------...........((((--------------((.(((((((.(((((....((((((((((((((.....)))))))))))).))..)))))....))))))).-...)))))). ( -37.20) >DroSim_CAF1 6849 99 + 1 G------ACACACACCGAGCUG--------------GUAUUUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCGAAAG-GACACCAGCC .------...........((((--------------((.(((((((.(((((....((((((((((((((.....)))))))))))).))..)))))....))))))).-...)))))). ( -37.20) >DroEre_CAF1 5125 83 + 1 ------------------------------------AUAUUUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCAGAAG-GACACCAGCC ------------------------------------....................((((((((((((((.....)))))))))))).))..(((((.((((.....))-))..))))). ( -29.50) >DroYak_CAF1 17279 99 + 1 G------ACACACACCAAGCUGC-------------AUAUUUUGAACUCAAUGAUGCGGCAGGCGGUCAACGCCUUCGACUGUCUGCACGAAGUUGGCUCCUUUAAAAG-GACACCAGC- (------(((........(((((-------------((...(((....)))..)))))))(((((.....))))).....))))........(((((.((((.....))-))..)))))- ( -32.50) >DroPer_CAF1 5628 119 + 1 GGGACACAUACACACAGAGCUGGACACUCUGCUGGGGUAUUUUGAAUUCAAUGAUGCAGCAAGCGGUCAACGCCUUUGACUGCUUGCAAGCAGCCGGCUCCUUCAAAAGGGACACCACC- ((..............(((((((.....(((((...((((((((....))).))))).((((((((((((.....)))))))))))).))))))))))))........((....)).))- ( -43.20) >consensus G______ACACACACCGAGCUG______________GUAUUUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCAAAAG_GACACCAGCC .......................................(((((((.(((((....((((((((((((((.....)))))))))))).))..)))))....)))))))............ (-23.31 = -23.40 + 0.09)


| Location | 22,639,681 – 22,639,791 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -22.29 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.773072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22639681 110 + 23771897 UUUAAAUUCAAUGAUGCGGCAGGCGGUAAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCGAAAG-GACGCCAACUCGGCCAGCUGGUGAAAAAAACC---------GGCGAGACC ......(((......((.(((((((((...........))))))))).(((.((((((((((.....))-)).)))))))))))..((((((.......)))---------))))))... ( -42.70) >DroPse_CAF1 5614 111 + 1 UUUGAAUUCAAUGAUGCAGCAAGCGGUCAACGCCUUUGACUGCUUGCAAGCAGCCGGCUCCUUCAAAAGGGACACCACC--GGGCA------CAAA-AAACCAGGCUCUCGGCCGAGGCA ...........((.(((.((((((((((((.....))))))))))))......((((.((((......)))).....))--)))))------))..-...((.(((.....)))..)).. ( -38.10) >DroSim_CAF1 6869 109 + 1 UUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCGAAAG-GACACCAGCCCGGCCAGCUGGUGAAA-AAACC---------GGCGAGACC ......(((.......((((((((((((((.....)))))))))))).))..(((((.((((.....))-))(((((((.......)))))))...-...))---------))))))... ( -41.90) >DroEre_CAF1 5129 109 + 1 UUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCAGAAG-GACACCAGCCCGGCCAGCUGGUGAAA-AAACA---------GCCGAGACC ................((((((((((((((.....)))))))))))).))...(((((((((.....))-))(((((((.......)))))))...-.....---------))))).... ( -41.70) >DroYak_CAF1 17300 108 + 1 UUUGAACUCAAUGAUGCGGCAGGCGGUCAACGCCUUCGACUGUCUGCACGAAGUUGGCUCCUUUAAAAG-GACACCAGC-CGGCCAGCUGGUGAAA-AAACU---------GCCGAGACC ......(((.......((((((((((((.........)))))))))).)).....(((((((.....))-))(((((((-......)))))))...-.....---------))))))... ( -41.50) >DroPer_CAF1 5668 111 + 1 UUUGAAUUCAAUGAUGCAGCAAGCGGUCAACGCCUUUGACUGCUUGCAAGCAGCCGGCUCCUUCAAAAGGGACACCACC--GGGGA------CAAA-AAACCAGGCUCUCGGCCGAGGCA ..............(((.((((((((((((.....))))))))))))..)))(((((.((((......))))..)).((--(((..------(...-.......)..)))))....))). ( -39.10) >consensus UUUGAAUUCAAUGAUGCGGCAGGCGGUCAACGCCUUUGACUGUCUGCACGAAGUUGGCUCCUUCAAAAG_GACACCAGC_CGGCCAGCUGGUGAAA_AAACC_________GCCGAGACC ......(((.........((((((((((((.....))))))))))))....((((((((......................)))))))).........................)))... (-22.29 = -23.65 + 1.36)


| Location | 22,639,721 – 22,639,812 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -26.38 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
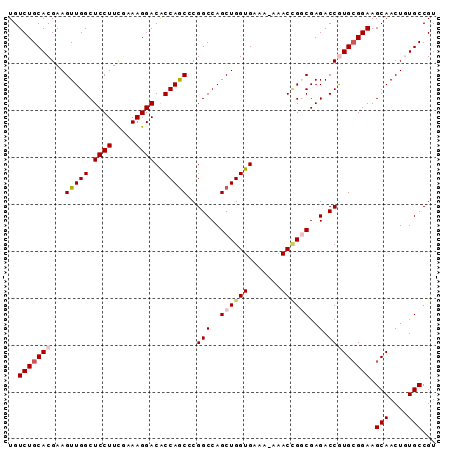
>3L_DroMel_CAF1 22639721 91 + 23771897 UGUCUGCACGAAGUUGGCUCCUUCGAAAGGACGCCAACUCGGCCAGCUGGUGAAAAAAACCGGCGAGACCGUGCGGAAGCAACUGUGCCGU ..(((((((((.((((((((((.....)))).))))))))(((..((((((.......))))))..).))))))))).(((....)))... ( -40.70) >DroSec_CAF1 8516 90 + 1 UGUCUGCACGAAGUUGGCUCCUUCGAAAGGACACCAGCCCGGCCAGCUGGUGAAA-AAACCGGCGAGACCGUGCGGAAGCAACUGUGCCGU ..(((((((...(((((.((((.....))))..)))))..(((..((((((....-..))))))..).))))))))).(((....)))... ( -34.80) >DroSim_CAF1 6909 90 + 1 UGUCUGCACGAAGUUGGCUCCUUCGAAAGGACACCAGCCCGGCCAGCUGGUGAAA-AAACCGGCGAGACCGUGCGGAAGCAACUGUGCCGU ..(((((((...(((((.((((.....))))..)))))..(((..((((((....-..))))))..).))))))))).(((....)))... ( -34.80) >DroEre_CAF1 5169 90 + 1 UGUCUGCACGAAGUUGGCUCCUUCAGAAGGACACCAGCCCGGCCAGCUGGUGAAA-AAACAGCCGAGACCAUGCGGAAGCAACUAUGCCGU ..((((((.(...(((((((((.....))))(((((((.......)))))))...-.....)))))...).)))))).(((....)))... ( -30.00) >DroYak_CAF1 17340 89 + 1 UGUCUGCACGAAGUUGGCUCCUUUAAAAGGACACCAGC-CGGCCAGCUGGUGAAA-AAACUGCCGAGACCAUGAGGAAGCAACUAUGCCGU .((.(((......(((((((((.....))))(((((((-......)))))))...-.....)))))..((....))..)))))........ ( -25.60) >consensus UGUCUGCACGAAGUUGGCUCCUUCGAAAGGACACCAGCCCGGCCAGCUGGUGAAA_AAACCGGCGAGACCGUGCGGAAGCAACUGUGCCGU ..(((((((...(((((.((((.....))))..)))))..(((..((((((.......))))))..).))))))))).(((....)))... (-26.38 = -27.30 + 0.92)

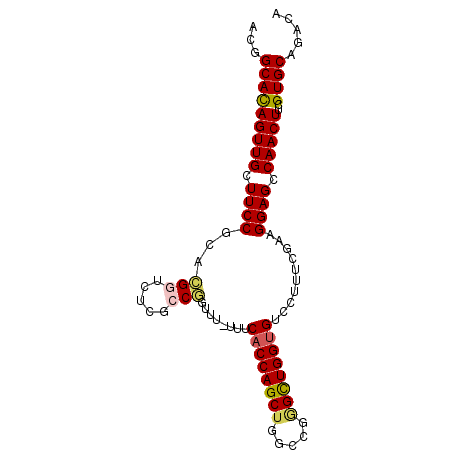
| Location | 22,639,721 – 22,639,812 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
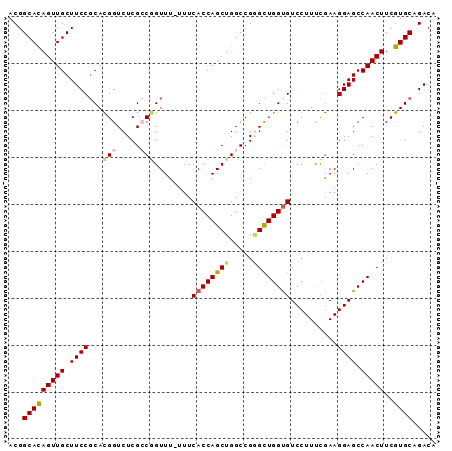
>3L_DroMel_CAF1 22639721 91 - 23771897 ACGGCACAGUUGCUUCCGCACGGUCUCGCCGGUUUUUUUCACCAGCUGGCCGAGUUGGCGUCCUUUCGAAGGAGCCAACUUCGUGCAGACA ..((((....))))((.((((((((..((.(((.......))).)).))))((((((((.((((.....)))))))))))).)))).)).. ( -35.70) >DroSec_CAF1 8516 90 - 1 ACGGCACAGUUGCUUCCGCACGGUCUCGCCGGUUU-UUUCACCAGCUGGCCGGGCUGGUGUCCUUUCGAAGGAGCCAACUUCGUGCAGACA ...((((((((((...(((.((((((.(((((((.-.......))))))).)))))))))((((.....))))).)))))..))))..... ( -33.40) >DroSim_CAF1 6909 90 - 1 ACGGCACAGUUGCUUCCGCACGGUCUCGCCGGUUU-UUUCACCAGCUGGCCGGGCUGGUGUCCUUUCGAAGGAGCCAACUUCGUGCAGACA ...((((((((((...(((.((((((.(((((((.-.......))))))).)))))))))((((.....))))).)))))..))))..... ( -33.40) >DroEre_CAF1 5169 90 - 1 ACGGCAUAGUUGCUUCCGCAUGGUCUCGGCUGUUU-UUUCACCAGCUGGCCGGGCUGGUGUCCUUCUGAAGGAGCCAACUUCGUGCAGACA ..((((....))))((.((((((....((((.(((-(..((((((((.....)))))))).......)))).))))....)))))).)).. ( -32.50) >DroYak_CAF1 17340 89 - 1 ACGGCAUAGUUGCUUCCUCAUGGUCUCGGCAGUUU-UUUCACCAGCUGGCCG-GCUGGUGUCCUUUUAAAGGAGCCAACUUCGUGCAGACA ..((((....))))..(((((((....(((.....-...((((((((....)-)))))))((((.....)))))))....))))).))... ( -29.80) >consensus ACGGCACAGUUGCUUCCGCACGGUCUCGCCGGUUU_UUUCACCAGCUGGCCGGGCUGGUGUCCUUUCGAAGGAGCCAACUUCGUGCAGACA ...(((((((((.((((...(((.....)))........((((((((.....))))))))..........)))).)))))..))))..... (-26.66 = -26.90 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:57 2006