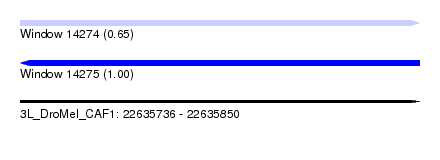
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,635,736 – 22,635,850 |
| Length | 114 |
| Max. P | 0.999595 |

| Location | 22,635,736 – 22,635,850 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -7.20 |
| Energy contribution | -8.52 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22635736 114 + 23771897 UCCCAUGCAGAAUUACGGAAAAAAUGUACAAUGUAAAAAAUGUGAA------AGGGUCAAUGCGAAUGUCUGAUACCCUCCACGCAUGUGUACGUAACUAGUAAGAAUUCUGCAUAGAAA ....((((((((((.........(((((((.........(((((..------(((((..(.((....)).)...)))))...))))).)))))))..........))))))))))..... ( -27.21) >DroSec_CAF1 4305 93 + 1 UCCCAUGCAGAAUUACGUAAAAAAUGUACAGUGUAAAAAACGU---------------------AAUGUCUGAUCCCCUCCACACAUAUGUGCAUAAC------GAGUUCCGCAUAGUAA ....((((.(((((.(((.....(((((((((.......))..---------------------.((((.((........)).)))).))))))).))------)))))).))))..... ( -18.60) >DroSim_CAF1 2653 93 + 1 UCCCAUGCAGAAUUACGGAAAAAAUGUACAGUGUAAAAAACGU---------------------AAUGUCUGAUCCCCUCCACACAUAUGUACAUAAC------GAAUUCCGCAUAGUAA ....((((.(((((.((......(((((((((.......))..---------------------.((((.((........)).)))).)))))))..)------)))))).))))..... ( -17.90) >DroEre_CAF1 983 111 + 1 UCCCAUGCAGAAUUACAAAAAAAAUGUACAAUGUAAAAAA-GUAAAAACCUAAGGGCCAAUGCGAAUGCCUGAUGCCCUGCACACAUAGAUG--------GUACGAACUCCGCAUAGCAA .....(((.....((((.......))))..((((......-...........(((((....((....)).....)))))............(--------(........)))))).))). ( -15.20) >DroYak_CAF1 13711 106 + 1 UCCCAUGCAGAAUUACGAAAAAAAUUUACAAUGUAAAAAAUGUAAC------AGGGCAAAUGCGAAUGCCGGAUCCCCUGCUCACAUAAAUG--------GUAAAAAUUCUGCAUAGAAC ....((((((((((..........(((((...)))))..((((..(------((((...((.((.....)).)).)))))...)))).....--------.....))))))))))..... ( -23.40) >consensus UCCCAUGCAGAAUUACGAAAAAAAUGUACAAUGUAAAAAACGU_A_______AGGG__AAUGCGAAUGUCUGAUCCCCUCCACACAUAUGUGC_UAAC__GUA_GAAUUCCGCAUAGAAA ....((((.(((((.........(((((((((((.................................................)))).)))))))..........))))).))))..... ( -7.20 = -8.52 + 1.32)



| Location | 22,635,736 – 22,635,850 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22635736 114 - 23771897 UUUCUAUGCAGAAUUCUUACUAGUUACGUACACAUGCGUGGAGGGUAUCAGACAUUCGCAUUGACCCU------UUCACAUUUUUUACAUUGUACAUUUUUUCCGUAAUUCUGCAUGGGA .((((((((((((((............(((((.(((.(((((((((....(.....)......)))).------)))))........))))))))...........)))))))))))))) ( -28.50) >DroSec_CAF1 4305 93 - 1 UUACUAUGCGGAACUC------GUUAUGCACAUAUGUGUGGAGGGGAUCAGACAUU---------------------ACGUUUUUUACACUGUACAUUUUUUACGUAAUUCUGCAUGGGA ...(((((((((((((------.((.(((((....))))))).)))........((---------------------((((.....................)))))))))))))))).. ( -23.80) >DroSim_CAF1 2653 93 - 1 UUACUAUGCGGAAUUC------GUUAUGUACAUAUGUGUGGAGGGGAUCAGACAUU---------------------ACGUUUUUUACACUGUACAUUUUUUCCGUAAUUCUGCAUGGGA ...(((((((((((((------(..(((((((...((((((((((...........---------------------...)))))))))))))))))......)).)))))))))))).. ( -28.64) >DroEre_CAF1 983 111 - 1 UUGCUAUGCGGAGUUCGUAC--------CAUCUAUGUGUGCAGGGCAUCAGGCAUUCGCAUUGGCCCUUAGGUUUUUAC-UUUUUUACAUUGUACAUUUUUUUUGUAAUUCUGCAUGGGA ...(((((((((((...(((--------.......(((((((((((.....((....))....))))..((((....))-))........))))))).......)))))))))))))).. ( -32.64) >DroYak_CAF1 13711 106 - 1 GUUCUAUGCAGAAUUUUUAC--------CAUUUAUGUGAGCAGGGGAUCCGGCAUUCGCAUUUGCCCU------GUUACAUUUUUUACAUUGUAAAUUUUUUUCGUAAUUCUGCAUGGGA .((((((((((((((.....--------.....((((((.(((((.(....((....))...).))))------)))))))..(((((...)))))..........)))))))))))))) ( -32.20) >consensus UUACUAUGCGGAAUUC_UAC__GUUA_GCACAUAUGUGUGGAGGGGAUCAGACAUUCGCAUU__CCCU_______U_ACAUUUUUUACAUUGUACAUUUUUUCCGUAAUUCUGCAUGGGA ...((((((((((....................((((.((........)).)))).............................((((................)))))))))))))).. (-14.95 = -14.79 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:54 2006