
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,594,286 – 22,594,376 |
| Length | 90 |
| Max. P | 0.594468 |

| Location | 22,594,286 – 22,594,376 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
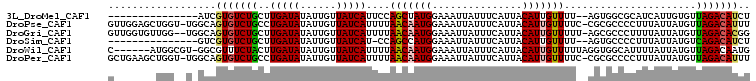
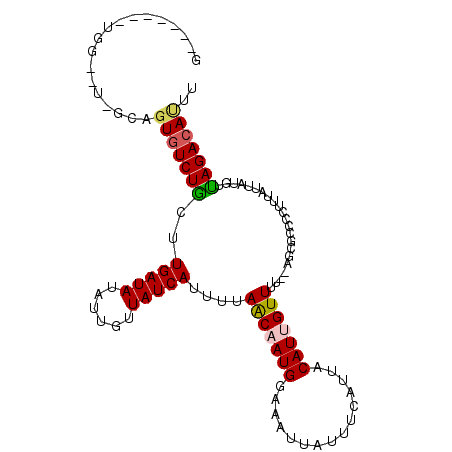
>3L_DroMel_CAF1 22594286 90 + 23771897 ---------------AUCGUGUCUGCUUGAUAUAUUGUUAUCAUUCCAGCUAUGGAAAUUAUUUCAUUACAUUGUUUU--AGUGGCGCAUCAUUGUGUUAGACAUCU ---------------...(((((((..(((((......)))))(((((....))))).......(((((........)--))))(((((....)))))))))))).. ( -19.00) >DroPse_CAF1 61464 105 + 1 GUUGGAGCUGGU-UGGCAGUGUCUGCCUGAUAUAUUGUUAUCAUUUUAACAAUGGAAAUUAUUUCAUUACAUUGUUUUC-CGCGCCCCUUUAUUAUGUUAGACAUUU ...((.(((((.-.((((((((.((..((((.((((((((......))))))))...))))...))..))))))))..)-)).)).))................... ( -22.70) >DroGri_CAF1 53782 104 + 1 GUUGGUGUUGG--UGGCAGUGUCUGCUUGAUAUAUUGUUAUCAUUUUAACAAUGGAAAUUAUUUCAUUACAUUGUUUUU-AGCGCCCUUUUAUUAUGUUAGACACGG ...((((((((--..(((((((.((..((((.((((((((......))))))))...))))...))..)))))))..))-))))))..................... ( -24.50) >DroSim_CAF1 51093 89 + 1 ---------------GUCGUGUCUGCUUGAUAUAUUGUUAUCAU-CCAGCCAUGGAAAUUAUUUCAUUACAUUGUUUU--AGUGCCCCUUUAUUAUGUCAGACAUCU ---------------((((((.(((..(((((......))))).-.))).)))((..((((...((......))...)--)))..)).............))).... ( -16.30) >DroWil_CAF1 68218 100 + 1 C------AUGGCGU-GGCGUUUCUACUUGAUAUAUUGUUAUCAUUUUAACAAUGGAAAUUAUUUCAUUACAUUGUUUUUAGGUGGCAUUUUAUUAUGUUAGACAAUG .------.......-...((((..((.(((((...(((((((.....(((((((...............)))))))....)))))))...))))).)).)))).... ( -17.96) >DroPer_CAF1 68629 105 + 1 GCUGAAGCUGGU-UGGCAGUGUCUGCCUGAUAUAUUGUUAUCAUUUUAACAAUGGAAAUUAUUUCAUUACAUUGUUUUC-CGCGCCCCUUUAUUAUGUUAGACAUUU ((..(......)-..))(((((((((.(((((.(((((((......)))))))(((((.................))))-).........))))).).)))))))). ( -21.03) >consensus G_______UGG__U_GCAGUGUCUGCUUGAUAUAUUGUUAUCAUUUUAACAAUGGAAAUUAUUUCAUUACAUUGUUUU__AGCGCCCCUUUAUUAUGUUAGACAUUU ..................(((((((..(((((......)))))....(((((((...............)))))))......................))))))).. (-12.84 = -12.89 + 0.06)

| Location | 22,594,286 – 22,594,376 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
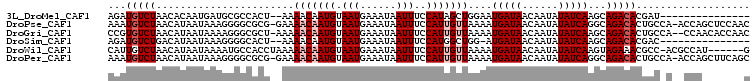
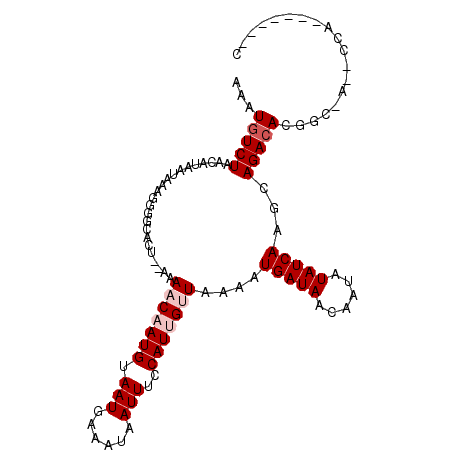
>3L_DroMel_CAF1 22594286 90 - 23771897 AGAUGUCUAACACAAUGAUGCGCCACU--AAAACAAUGUAAUGAAAUAAUUUCCAUAGCUGGAAUGAUAACAAUAUAUCAAGCAGACACGAU--------------- ...(((((..(.....)..((......--.....................(((((....)))))(((((......))))).)))))))....--------------- ( -11.30) >DroPse_CAF1 61464 105 - 1 AAAUGUCUAACAUAAUAAAGGGGCGCG-GAAAACAAUGUAAUGAAAUAAUUUCCAUUGUUAAAAUGAUAACAAUAUAUCAGGCAGACACUGCCA-ACCAGCUCCAAC ...................(((((..(-(..(((((((.(((......)))..)))))))....(((((......)))))(((((...))))).-.)).)))))... ( -26.30) >DroGri_CAF1 53782 104 - 1 CCGUGUCUAACAUAAUAAAAGGGCGCU-AAAAACAAUGUAAUGAAAUAAUUUCCAUUGUUAAAAUGAUAACAAUAUAUCAAGCAGACACUGCCA--CCAACACCAAC ..((((((..(.........)...(((-...(((((((.(((......)))..)))))))....(((((......)))))))))))))).....--........... ( -16.40) >DroSim_CAF1 51093 89 - 1 AGAUGUCUGACAUAAUAAAGGGGCACU--AAAACAAUGUAAUGAAAUAAUUUCCAUGGCUGG-AUGAUAACAAUAUAUCAAGCAGACACGAC--------------- ...((((((((((......((....))--......))))............((((....)))-)(((((......)))))..))))))....--------------- ( -13.50) >DroWil_CAF1 68218 100 - 1 CAUUGUCUAACAUAAUAAAAUGCCACCUAAAAACAAUGUAAUGAAAUAAUUUCCAUUGUUAAAAUGAUAACAAUAUAUCAAGUAGAAACGCC-ACGCCAU------G .....((((......................(((((((.(((......)))..)))))))....(((((......)))))..))))......-.......------. ( -9.90) >DroPer_CAF1 68629 105 - 1 AAAUGUCUAACAUAAUAAAGGGGCGCG-GAAAACAAUGUAAUGAAAUAAUUUCCAUUGUUAAAAUGAUAACAAUAUAUCAGGCAGACACUGCCA-ACCAGCUUCAGC ...................(((((..(-(..(((((((.(((......)))..)))))))....(((((......)))))(((((...))))).-.)).)))))... ( -23.50) >consensus AAAUGUCUAACAUAAUAAAGGGGCACU__AAAACAAUGUAAUGAAAUAAUUUCCAUUGUUAAAAUGAUAACAAUAUAUCAAGCAGACACGGC_A__CCA_______C ...(((((.......................(((((((.(((......)))..)))))))....(((((......)))))...)))))................... ( -9.07 = -9.90 + 0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:41 2006