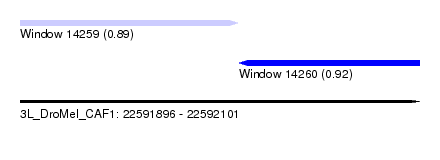
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,591,896 – 22,592,101 |
| Length | 205 |
| Max. P | 0.923690 |

| Location | 22,591,896 – 22,592,008 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -15.63 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
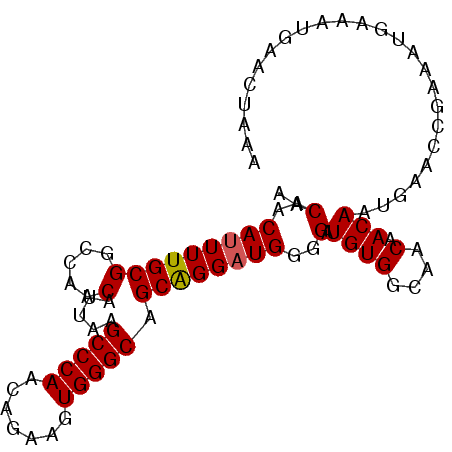
>3L_DroMel_CAF1 22591896 112 + 23771897 CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUACUUUU-CACCACAAUCGUU-UUCCAUUGUUGUAAUACAAUUUUCUUGUCGCAUUUCGGCCAAGUUAUUCUAUGCCACUCUAU ................((((((....((((((.....-.((.(((((.(..-..).))))).))..)))))).....)))))).....(((..((.....))..)))....... ( -19.60) >DroVir_CAF1 61305 110 + 1 CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUACUUUU-CGCCGCAAUCGCU-UUCCAUUGUUGAAAUACAAUUUUCUUGUCGCAUUUUAGUCAAGUUAUUUUAUUUUAUUCU-- .....((.((((..(.((((((....((((((..(((-((..(((((....-....)))))))))))))))).....)))))))..)))).))...................-- ( -21.70) >DroGri_CAF1 50736 110 + 1 CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUACUUUU-CGCCGCAAUCUCU-UUCCAUUGUUGUAAUACAAUUUUCUUGUCGCAUUUUAGUCAAGUUAUUUUAUUUUAUUGU-- .....((.((((..(.((((((....((((((....(-..(.(((((....-....))))).)..))))))).....)))))))..)))).))...................-- ( -19.50) >DroWil_CAF1 65104 114 + 1 CUUCAUGUCUCAUUUAGCGACAUUUCAUUGUACUUUUGCACCGCAAUCGCUUUUCCAUUGUUGUAAUACAAUUUUCUUGUCGCAUUUUAGUCAAGUUAUUUUAUUUUAUUUUCU ................((((((....((((((...(((((..(((((.(.....).)))))))))))))))).....))))))............................... ( -17.30) >DroMoj_CAF1 76567 110 + 1 CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUAGUUUU-CGCCGCAAUCGCU-UUCCAUUGUUGAAAUACAAUUUUCUUGUCGCAUUUUAGUCAAGUUAUUUUAUUUUAUUGU-- .....((.((((..(.((((((....((((((..(((-((..(((((....-....)))))))))))))))).....)))))))..)))).))...................-- ( -21.70) >DroAna_CAF1 43553 112 + 1 CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUACUUUU-CACCACAAUCGCU-UUCCAUUGUUGUAAUACAAUUUUCUUGUCGCAUUUUGGCCAAGUUAUUCUAUUCCACUCUGU .....((.((((..(.((((((....((((((.....-.((.(((((....-....))))).))..)))))).....)))))))..)))).))..................... ( -17.40) >consensus CUUCAUGUCUAAUUUAGCGACAUUUUAUUGUACUUUU_CACCGCAAUCGCU_UUCCAUUGUUGUAAUACAAUUUUCUUGUCGCAUUUUAGUCAAGUUAUUUUAUUUUAUUCU__ .....((.((((..(.((((((....((((((.......((.(((((.........))))).))..)))))).....)))))))..)))).))..................... (-15.63 = -15.77 + 0.14)



| Location | 22,592,008 – 22,592,101 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22592008 93 - 23771897 AACAACAUUUUGCGGCCAACAUUAAGCCCAACAGAAGUGGGCAGCAGGAUGGUGAUGUGGCAACAACAAUGAACCGAAAUGAAAUGAACUAAA .....(((((((..((((.((((..(((((.......))))).((......))))))))))..))).))))...................... ( -21.40) >DroSec_CAF1 48707 93 - 1 AACAACAUUUUGCGGCCAACAUAAGGCCCAACAGAAGUGGGCAGCAGGAUGGGGAUGUGGCAACAACAAUGAACCGAAAUGAAAUGAACUAAA .....((((((.((((((.(.....(((((.......))))).....).)))...((((....).))).....)))....))))))....... ( -21.50) >DroSim_CAF1 48801 93 - 1 AACAACAUUUUGCGGCCAACAUUAAGCCCAACAGAAGUGGGCAGCGGGAUGGGGAUGUGGCAGCAACAAUGAACCGAAAUGAAAUGAACUAAA .....((((((((.((((.((((..(((((.......)))))..(......).)))))))).)))).))))...................... ( -22.60) >DroEre_CAF1 49404 92 - 1 AACAACAUUUUGCGGCCAACAUUAAGCCCAACAGAAGUGGGCAGCAGGCUGGGGAUGUGGCAGCAACAAAGAGC-GAAAUGAAAUGAACUAAA .....((((((((.((((.((((..(((((.......)))))((....))...))))))))...........))-))))))............ ( -24.70) >DroYak_CAF1 62128 93 - 1 AACAACAUUUUGCGGCCAACAUUAAGCCCAACAGAAGUGGGCAGCAGGAUGGGGAUGUGGCAACAACAAUGAGCCGAAAUGAAAUGAACUAAA .....((((((.((((((.((((..(((((.......))))).....))))....((((....).))).)).))))....))))))....... ( -23.70) >consensus AACAACAUUUUGCGGCCAACAUUAAGCCCAACAGAAGUGGGCAGCAGGAUGGGGAUGUGGCAACAACAAUGAACCGAAAUGAAAUGAACUAAA ..(..(((((((((.....).....(((((.......))))).))))))))..).((((....).)))......................... (-18.46 = -18.50 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:39 2006