| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,534,933 – 22,535,049 |
| Length | 116 |
| Max. P | 0.975373 |

| Location | 22,534,933 – 22,535,031 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22534933 98 - 23771897 GUCGUGCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAAC--------CAGAC---GACUGGCUG--GA------UCCCC--ACCACUCCU- ........(((.((.(((((((((..(.....((((((((....)).)))))).)..))))))))).)--------).)))---((.(((.((--(.------...))--)))).))..- ( -36.80) >DroPse_CAF1 42343 117 - 1 GUCGUCCAGU---ACUGCAUGUUGUGGCAUGCGGUUGUGCAACGGCAACAACUUUUGCAACAUGCAACCAACGAACCAGACGAGCUCUGGCUCCCGCUCCCGCUCCCCCUCCCGCUCCUG (((.((....---..(((((((((..(.....((((((((....)).)))))).)..)))))))))......))....)))((((...(((....)))...))))............... ( -32.60) >DroSec_CAF1 18436 101 - 1 GUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAAC--------CAGACGACGACUGGCUG--GA------UCCCC--GCCACUCCU- ((((((..(((.((.(((((((((..(.....((((((((....)).)))))).)..))))))))).)--------).)))))))))((((.(--(.------...))--)))).....- ( -44.70) >DroSim_CAF1 19553 101 - 1 GUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAAC--------CAGACGACGACUGGCUG--GA------UCCCC--GCCACUCCU- ((((((..(((.((.(((((((((..(.....((((((((....)).)))))).)..))))))))).)--------).)))))))))((((.(--(.------...))--)))).....- ( -44.70) >DroEre_CAF1 17838 98 - 1 GUCGUCCGGCCAGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAAC--------CAGAC---GACUGGCCG--GA------UCCCC--UCCACUUCU- ...((((((((.((.(((((((((..(.....((((((((....)).)))))).)..))))))))).)--------)((..---..)))))))--))------)....--.........- ( -40.30) >DroAna_CAF1 47863 101 - 1 GUCGUCCAGCCAGGCUGCAUGUUGCGGCAUGCGGUUGUGCAACGGCAACAACUUCUGCUACAUGCAAC--------CAGAC---GACUUCCUC--GA---CACUCCCC--GCCCGUCCG- ((((((......((.(((((((.((((.....((((((((....)).)))))).)))).))))))).)--------).)))---)))......--((---(.......--....)))..- ( -32.00) >consensus GUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAAC________CAGAC___GACUGGCUG__GA______UCCCC__GCCACUCCU_ ((((((......)))((((((((((((.....((((((((....)).)))))).))))))))))))............)))....................................... (-24.60 = -24.68 + 0.08)

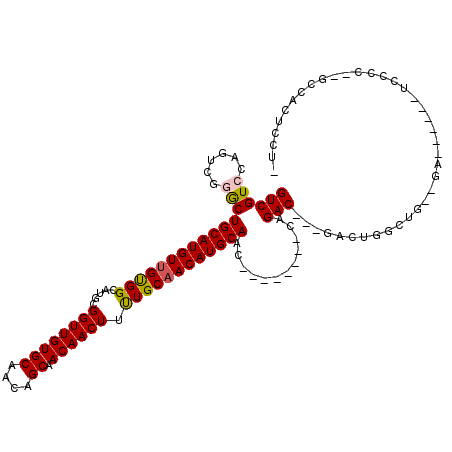

| Location | 22,534,959 – 22,535,049 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22534959 90 + 23771897 UCUGGUUGCAUGUUGCAAAAGUUGUUGCUGUUGCACAACCGCAUGCCACAACAUGCAGCCCGACUGCACGACCUGGGGGACUCGGGUUAC ((.((((((((((((((...(((((.((....)))))))....)))...))))))))))).))......(((((((....).)))))).. ( -36.30) >DroSec_CAF1 18465 90 + 1 UCUGGUUGCAUGUUGCAAAAGUUGUUGCUGUUGCACAACCGCAUGCCACAACAUGCAGCCCGACUGGACGACCUGGGGGACUGGGGUUAC ((.((((((((((((((...(((((.((....)))))))....)))...))))))))))).))........((..(....)..))..... ( -35.30) >DroSim_CAF1 19582 90 + 1 UCUGGUUGCAUGUUGCAAAAGUUGUUGCUGUUGCACAACCGCAUGCCACAACAUGCAGCCCGACUGGACGACCUGGGGGACUGGGGUUAC ((.((((((((((((((...(((((.((....)))))))....)))...))))))))))).))........((..(....)..))..... ( -35.30) >DroEre_CAF1 17864 90 + 1 UCUGGUUGCAUGUUGCAAAAGUUGUUGCUGUUGCACAACCGCAUGCCACAACAUGCAGCCUGGCCGGACGACCUGGGGGAGUGGGCUUAC ...((((((((((((((...(((((.((....)))))))....)))...))))))))))).((((.(.(..((...))..)).))))... ( -33.00) >DroAna_CAF1 47892 86 + 1 UCUGGUUGCAUGUAGCAGAAGUUGUUGCCGUUGCACAACCGCAUGCCGCAACAUGCAGCCUGGCUGGACGACCUGUGGGGCAAGAA---- ((((((((((.(((((((...)))))))...))))..)))(((((......))))).((((.((.((....)).)).)))).))).---- ( -30.10) >consensus UCUGGUUGCAUGUUGCAAAAGUUGUUGCUGUUGCACAACCGCAUGCCACAACAUGCAGCCCGACUGGACGACCUGGGGGACUGGGGUUAC ((.((((((((((((((...(((((.((....)))))))....)))...))))))))))).))........(((((....)))))..... (-29.00 = -29.84 + 0.84)

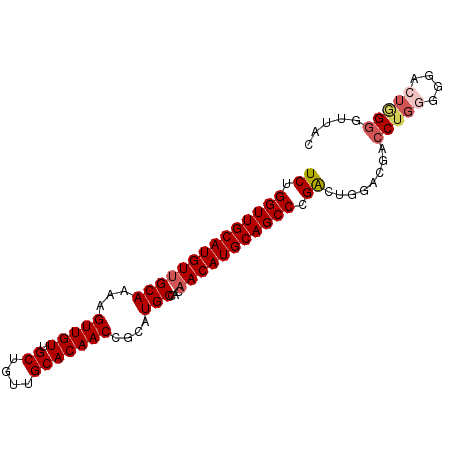

| Location | 22,534,959 – 22,535,049 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22534959 90 - 23771897 GUAACCCGAGUCCCCCAGGUCGUGCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAACCAGA ....((((((.((....)).)......))))).(((((((((..(.....((((((((....)).)))))).)..)))))))))...... ( -28.80) >DroSec_CAF1 18465 90 - 1 GUAACCCCAGUCCCCCAGGUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAACCAGA .................(((.((((....))))(((((((((..(.....((((((((....)).)))))).)..))))))))))))... ( -29.40) >DroSim_CAF1 19582 90 - 1 GUAACCCCAGUCCCCCAGGUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAACCAGA .................(((.((((....))))(((((((((..(.....((((((((....)).)))))).)..))))))))))))... ( -29.40) >DroEre_CAF1 17864 90 - 1 GUAAGCCCACUCCCCCAGGUCGUCCGGCCAGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAACCAGA .................((((....)))).((.(((((((((..(.....((((((((....)).)))))).)..))))))))).))... ( -31.30) >DroAna_CAF1 47892 86 - 1 ----UUCUUGCCCCACAGGUCGUCCAGCCAGGCUGCAUGUUGCGGCAUGCGGUUGUGCAACGGCAACAACUUCUGCUACAUGCAACCAGA ----.............(((......))).((.(((((((.((((.....((((((((....)).)))))).)))).))))))).))... ( -27.70) >consensus GUAACCCCAGUCCCCCAGGUCGUCCAGUCGGGCUGCAUGUUGUGGCAUGCGGUUGUGCAACAGCAACAACUUUUGCAACAUGCAACCAGA .................(((.((((....))))((((((((((((.....((((((((....)).)))))).)))))))))))))))... (-25.20 = -25.68 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:28 2006