
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,534,685 – 22,534,816 |
| Length | 131 |
| Max. P | 0.827041 |

| Location | 22,534,685 – 22,534,794 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -27.61 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
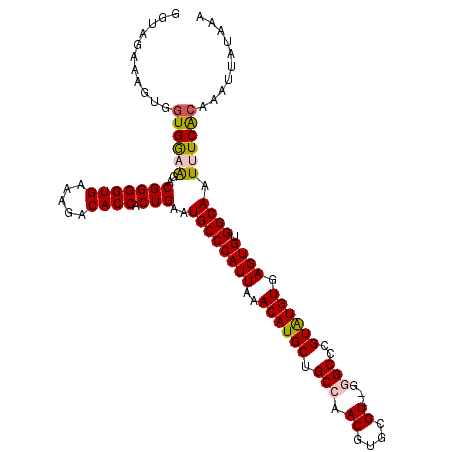
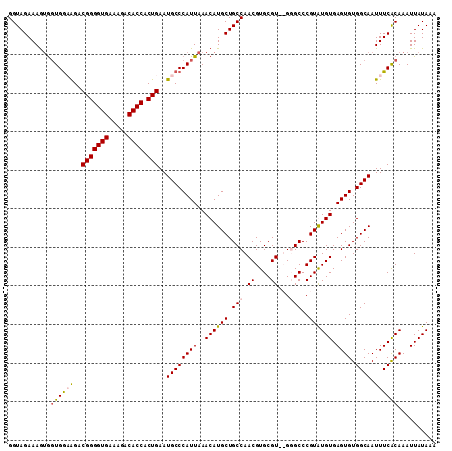
>3L_DroMel_CAF1 22534685 109 - 23771897 GGUAGAAAAUAGUGAAGGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU--GGGCCCGUAUGUGAGUGUGGCAAUUUCACAAAUUAUAAA ...........((((((..(((((((.....)))).)))..((((((((..((((((.(((.((....))--.)))..)))))).)))).)))).)))))).......... ( -34.50) >DroSec_CAF1 18186 111 - 1 GGUAGAAAGUGGUGGCAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGUAUGUGCCCGUGUGUGAGUGUGGCAAUUUCACAAAUUAUAAA (((((....((((((((..(((((((.....)))).)))..)).))))))......))))).(((.(((....))).))).((((((.........))))))......... ( -32.60) >DroSim_CAF1 19303 111 - 1 GGUAGAAAGUGGUGGCAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGUAUGUGCCCGUGUGUGAGUGUGGCAAUUUCACAAAUUAUAAA (((((....((((((((..(((((((.....)))).)))..)).))))))......))))).(((.(((....))).))).((((((.........))))))......... ( -32.60) >DroEre_CAF1 17605 108 - 1 GGUAGAAAGAGCUGGAAGUCGGGGUGGAA-ACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU--GGGCCCGUAUGUGAGUGUGGCAAUUUCGCAAAUUAUAAA ..........((.((((.((((((((...-.)))).)))).((((((((..((((((.(((.((....))--.)))..)))))).)))).)))).)))))).......... ( -37.60) >consensus GGUAGAAAGUGGUGGAAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU__GGGCCCGUAUGUGAGUGUGGCAAUUUCACAAAUUAUAAA ...........((((((..(((((((.....)))).)))..((((((((..((((((.(((.((....))...)))..)))))).)))).)))).)))))).......... (-27.61 = -28.05 + 0.44)

| Location | 22,534,724 – 22,534,816 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.79 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -20.70 |
| Energy contribution | -23.07 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
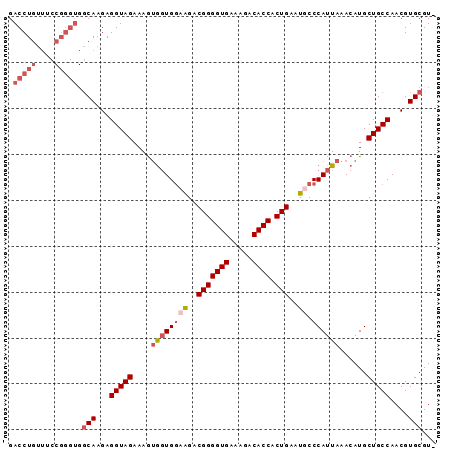
>3L_DroMel_CAF1 22534724 92 - 23771897 GACCUGUUUCCGGGUGGCAAGAGGUAGAAAAUAGUGAAGGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU- .(((((....))))).(((...(((((....(((((..((.(((((((.....)))).)))....)))))))......)))))....)))..- ( -25.50) >DroSec_CAF1 18226 93 - 1 GACCUGUUUCCGGGUGGCAAGAGGUAGAAAGUGGUGGCAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGUA .(((((....))))).(((...(((((....((((((((..(((((((.....)))).)))..)).))))))......)))))....)))... ( -32.00) >DroSim_CAF1 19343 93 - 1 GACCUGUUUCCGGGUGGCAAGAGGUAGAAAGUGGUGGCAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGUA .(((((....))))).(((...(((((....((((((((..(((((((.....)))).)))..)).))))))......)))))....)))... ( -32.00) >DroEre_CAF1 17644 81 - 1 GACCUG----------CCAAGAGGUAGAAAGAGCUGGAAGUCGGGGUGGAA-ACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU- ...(((----------((....)))))........((...((((((((...-.)))).))))...))........((((.........))))- ( -24.20) >consensus GACCUGUUUCCGGGUGGCAAGAGGUAGAAAGUGGUGGAAGACGGGGUGAAAGACACCACUGAAUGCCCAUUAAACAUGCUGCCAACGUGCGU_ .(((((....))))).(((...(((((....((((((((..(((((((.....)))).)))..)).))))))......)))))....)))... (-20.70 = -23.07 + 2.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:26 2006