| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,531,592 – 22,531,685 |
| Length | 93 |
| Max. P | 0.811262 |

| Location | 22,531,592 – 22,531,685 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
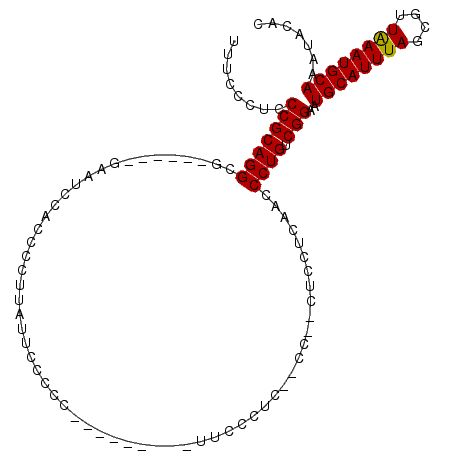
| Structure conservation index | 0.62 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
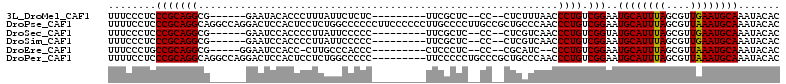
>3L_DroMel_CAF1 22531592 93 + 23771897 UUUCCCUCCCGCAGGCG------GAAUACACCCUUUAUUCUCUC---------UUCGCUC--CC--CUCUUUAACCCUGUCGGAAUGCAUUUAGCGUUGAAUGCAAAUACAC ......(((.(((((.(------(((((.......))))))...---------.......--..--.........))))).))).((((((((....))))))))....... ( -16.75) >DroPse_CAF1 38249 112 + 1 UUUUCCUCCCGCAGGCAGGCCAGGACUCCACUCCUCUGGCCCCCCUUCCCCCCUUGCCCCUUGCCGCUGCCCAACCCUGUCGGAAUGCAUUUAGCGUUAAAUGCAAAUACAC ......(((.(((((..(((((((..........)))))))..............((........))........))))).))).((((((((....))))))))....... ( -28.30) >DroSec_CAF1 15062 93 + 1 UUUCCCUCCCGCAGGCG------GAAUCCACCCCUUAUUCCCCC---------UUCGCUC--CC--CUCGUCAACCCUGUCGGUAUGCAUUUAGCGUUGAAUGCAAAUACAC .........((.(((.(------((((.........))))).))---------).))...--..--..((.((....)).))(((((((((((....))))))))..))).. ( -17.20) >DroSim_CAF1 15918 93 + 1 UUUCCCUCCCGCAGGCG------GAAUCCACCCCUUAUUCCCCC---------UUCGCUC--CC--CUCGUCAACCCUGUCGGAAUGCAUUUAGCGUUGAAUGCAAAUACAC ......(((.(((((.(------((((.........)))))...---------...((..--..--...))....))))).))).((((((((....))))))))....... ( -18.10) >DroEre_CAF1 14032 91 + 1 UUUCCCUGCCGCAGGCG-----GGAAUCCACC-CUUGCCCACCC---------CUCCCUC--CC--CGCAUC--CCCUGUCGGAAUGCAUUUAGCGUUAAAUGCAAAUACAC ........(((((((.(-----((......))-).(((......---------.......--..--.)))..--.)))).)))..((((((((....))))))))....... ( -20.29) >DroPer_CAF1 43043 103 + 1 UUUUCCUCCCGCAGGCAGGCCAGGACUCCACUCCUCUGGCCCCC---------UUCCCCCUGCCCGCUGCCCAACCCUGUCGGAAUGCAUUUAGCGUUAAAUGCAAAUACAC ......(((.(((((..(((((((..........)))))))...---------........((.....)).....))))).))).((((((((....))))))))....... ( -27.80) >consensus UUUCCCUCCCGCAGGCG______GAAUCCACCCCUUAUUCCCCC_________UUCCCUC__CC__CUCCUCAACCCUGUCGGAAUGCAUUUAGCGUUAAAUGCAAAUACAC ........(((((((............................................................)))).)))..((((((((....))))))))....... (-13.27 = -13.02 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:20 2006