
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,527,479 – 22,527,577 |
| Length | 98 |
| Max. P | 0.716536 |

| Location | 22,527,479 – 22,527,577 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.32 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
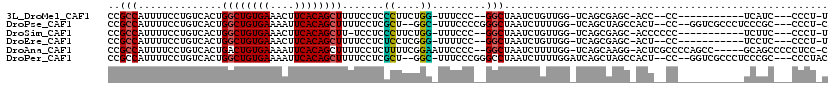
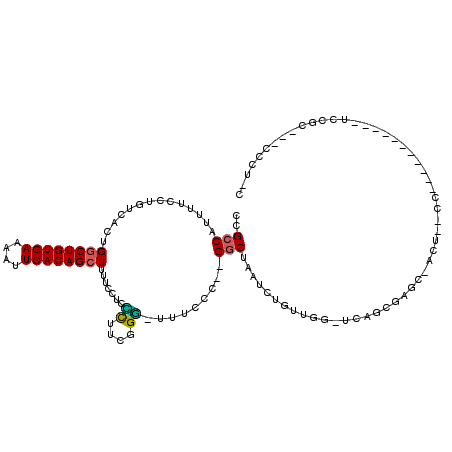
>3L_DroMel_CAF1 22527479 98 - 23771897 CCGCCAUUUUCCUGUCACUGGCUGUGAAACUUCACAGCUUUUCCUCCCUUCUGG-UUUCCC--GGCUAAUCUGUUGG-UCAGCGAGC-ACC--CC-----------UCAUC---CCCU-U ..(((..............((((((((....))))))))..............(-((..((--(((......)))))-..)))).))-...--..-----------.....---....-. ( -21.40) >DroPse_CAF1 33249 108 - 1 CCGCCAUUUUCCUGUCACUGGCUGUGAAAAUUCACAGCUUUUCCUCGCU--GGC-UUUCCCCGGGCUAAUCUUUUGG-UCAGCUAGCCACU--CC--GGUCGCCCUCCCGC---CCCU-C .............((.(((((((((((....)))))))........(((--(((-(.......((((((....))))-)))))))))....--.)--))).))........---....-. ( -28.11) >DroSim_CAF1 11874 99 - 1 CCGCCAUUUUCCUGUCACUGGCUGUGAAACUUCACAGCUU-UCCUCCCUUCUGG-UUUCCC--GGCUAAUCUGUUGG-UCAGCGAGC-ACCCCCC-----------UCUUC---CCCU-U ..(((..............((((((((....)))))))).-............(-((..((--(((......)))))-..)))).))-.......-----------.....---....-. ( -21.40) >DroEre_CAF1 9979 98 - 1 CCGCCAUUUUCCUGUCACUGGCUGUGAAACUUCACAGCUUUUCCUCUCCUCGGG-UUUUCC--GGCUAAUCUGUUGG-UCAGCGAGC-ACU--CC-----------UCCUC---CCCU-U ..(((.....((((.....((((((((....))))))))...........))))-......--))).........((-..((.(((.-...--.)-----------)))).---.)).-. ( -23.19) >DroAna_CAF1 41244 110 - 1 CCGCCAUUUUCCUGUCACUGACUGUGAAAAUUCACAGCUUUUCCUCUUUUCGGAAUUCCCC--GGCUAAUCUUUUGG-UCAGCAAGG-ACUCGCCCCAGCC-----GCAGCCCCCUCC-C ...........((((..(((.((((((....))))))....((((.....(((......))--)(((.(((....))-).))).)))-).......)))..-----))))........-. ( -24.30) >DroPer_CAF1 33635 110 - 1 CCGCCAUUUUCCUGUCACUGGCUGUGAAAAUUCACAGCUUUUCCUCGCU--GGC-UUUCCCGGGCCUAAUCUUUUGGAUCAGCUAGCCACU--CC--GGUCGCCCUCCCGC---CCCUAC .............((.(((((((((((....)))))))........(((--(((-(...(((((........)))))...)))))))....--.)--))).))........---...... ( -27.30) >consensus CCGCCAUUUUCCUGUCACUGGCUGUGAAAAUUCACAGCUUUUCCUCCCUUCGGG_UUUCCC__GGCUAAUCUGUUGG_UCAGCGAGC_ACU__CC___________UCCGC___CCCU_C ..(((..............((((((((....)))))))).......((....)).........)))...................................................... (-13.76 = -13.32 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:15 2006