
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
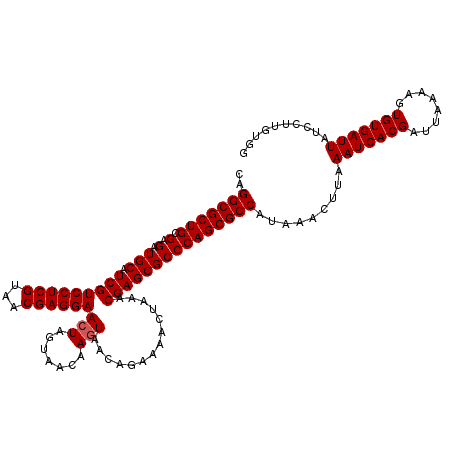
| Location | 22,510,698 – 22,510,866 |
| Length | 168 |
| Max. P | 0.996964 |

| Location | 22,510,698 – 22,510,805 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -31.35 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22510698 107 - 23771897 CAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAA------------CGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGG ..(((((((.(.(.(((.(((((((((...))))))(((.......)))..------------))))))))))))))).........(((((((........))))))).......... ( -32.80) >DroSec_CAF1 18756 119 - 1 CAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACGGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGG ..(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))).........(((((((........))))))).......... ( -33.10) >DroSim_CAF1 19194 119 - 1 CAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGG ..(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))).........(((((((........))))))).......... ( -33.10) >DroEre_CAF1 20835 119 - 1 CAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAAGUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGG ..(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))).........(((((((........))))))).......... ( -33.10) >consensus CAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCAUAAACUUAAAUCACGAUUAAAAGUGUGAUUAUCCUUGUGG ..(((((((.(.(.(((.(((((((((...))))))(((.......)))..............))))))))))))))).........(((((((........))))))).......... (-31.35 = -31.60 + 0.25)

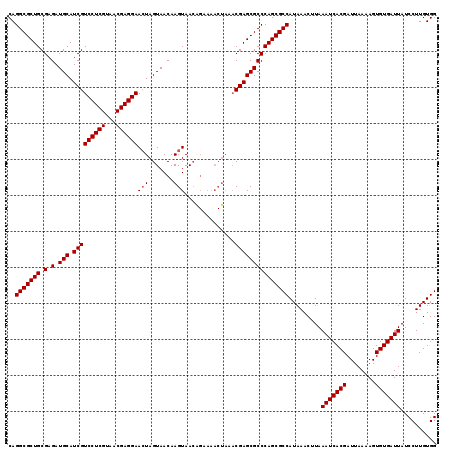
| Location | 22,510,738 – 22,510,844 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -35.32 |
| Energy contribution | -35.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22510738 106 - 23771897 GUCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAA------------CGAGCGCCCAGCGCCA .........(((((((.................))))))).(((((((.(.(.(((.(((((((((...))))))(((.......)))..------------))))))))))))))). ( -37.43) >DroSec_CAF1 18796 118 - 1 UCCGAGAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACGGAAAACUAAACGAGCGCCCAGCGCCA .........(((((((.................))))))).(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))). ( -37.73) >DroSim_CAF1 19234 118 - 1 UCCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCA .........(((((((.................))))))).(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))). ( -37.73) >DroEre_CAF1 20875 118 - 1 UCCGAUGGAGCCGUGAGAAAGGAAUUGAAAAAAUCGUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAAGUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCA (((...)))(((((((.................))))))).(((((((.(.(.(((.(((((((((...))))))..........(((........)))...))))))))))))))). ( -34.63) >consensus UCCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUAGUAACAAGUAACAGAAAACUAAACGAGCGCCCAGCGCCA .........(((((((.................))))))).(((((((.(.(.(((.(((((((((...))))))(((.......)))..............))))))))))))))). (-35.32 = -35.38 + 0.06)


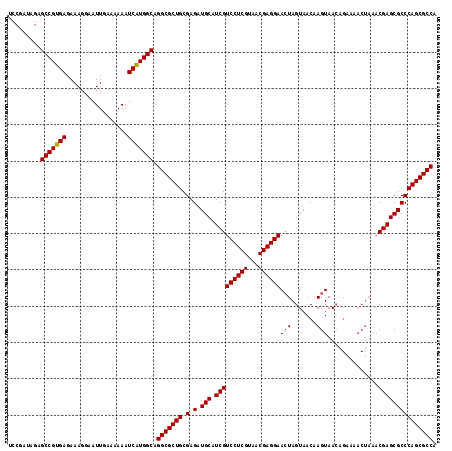
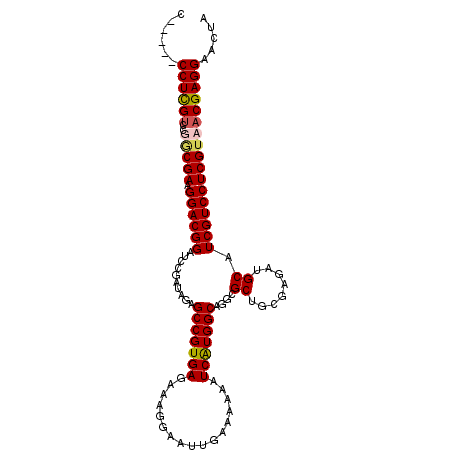
| Location | 22,510,765 – 22,510,866 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -32.23 |
| Energy contribution | -32.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22510765 101 - 23771897 C-----CCUUGACCGACGAAGGACGGAGUCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUA .-----(((((....((((.((((((.(((......(((((((.................))))))).)))((........)).))))))))))..)))))..... ( -32.13) >DroSec_CAF1 18835 101 - 1 C-----CCUCGUCCUGCGAAGGACGGAUCCGAGAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUA .-----((((((..(((((.(((((((((((((.(.(((((((.................)))))))...).)).)).)))...)))))))))))))))))..... ( -36.83) >DroSim_CAF1 19273 101 - 1 C-----CCUCGUCCUGCGAAGGACGGAUCCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUA .-----((((((..(((((.(((((((((((.(((.(((((((.................))))))).....))))).)))...)))))))))))))))))..... ( -36.53) >DroEre_CAF1 20914 106 - 1 CUCGUUCCUCGUCCGCCGAAGGACGGAUCCGAUGGAGCCGUGAGAAAGGAAUUGAAAAAAUCGUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAAGUA ....((((((((....(((.((((((...((((....((........)).))))......(((..((....))..)))......)))))))))..))))))))... ( -35.80) >consensus C_____CCUCGUCCGGCGAAGGACGGAUCCGAUAGAGCCGUGAGAAAGGAAUUGAAAAAAUCAUGGCAGGCGCUGCGAGAUGCAUCGUCCUCGUAACGAGGAACUA ......((((((..(((((.((((((..........(((((((.................)))))))....((........)).)))))))))))))))))..... (-32.23 = -32.73 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:10 2006