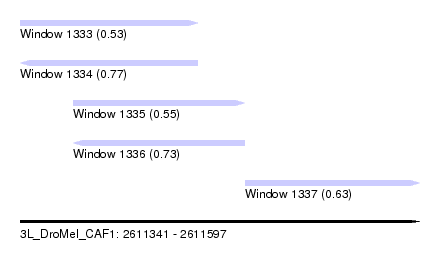
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,611,341 – 2,611,597 |
| Length | 256 |
| Max. P | 0.773274 |

| Location | 2,611,341 – 2,611,455 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -14.78 |
| Energy contribution | -19.38 |
| Covariance contribution | 4.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2611341 114 + 23771897 ----AAAUG--AAAUCCAUUUAUUUGGGGACUCCAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUUUACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAUCCAC ----(((((--.....)))))((((..(((((((...................((((.(((......)))..)))).((((((((((.....)))))))))).)))))))..)))).... ( -35.30) >DroSec_CAF1 9525 106 + 1 AGUUAAAUAAUAAAUCCAUUUAUUUGGGGACUCCAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU--------------AUUUAUCCACCUGUCAGCAGUUUUAAAAUCCAC ...............(((......)))(((....(((((...((((((.((.(((((.(((......))).))--------------)))....)).))))))..))))).....))).. ( -18.70) >DroSim_CAF1 12210 106 + 1 AGUUAAAUAAUAAAUCCAUUUAUUUGGGGACUCCAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU--------------AUUUAUCCACCUGUCAGCAGUUUUAAAAUCCAC ...............(((......)))(((....(((((...((((((.((.(((((.(((......))).))--------------)))....)).))))))..))))).....))).. ( -18.70) >DroEre_CAF1 9736 116 + 1 AAUGAAAUAAUAAAUCCUCAUAUUUGGGGACCCCAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUCAACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAA---- ..((((((......((((......((((((..((((......((.(....)))....))))..)).)))).......((((((((((.....))))))))))))))))))))....---- ( -33.00) >DroYak_CAF1 13002 120 + 1 AAUGAAAUAAUAAAUCCAUGUAUUUGGGGACCCAAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUCAACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUGAAAUCCGC ...............(((......)))(((..((((((((.............((...(((......)))....)).((((((((((.....))))))))))..))))))))...))).. ( -33.40) >consensus AAUGAAAUAAUAAAUCCAUUUAUUUGGGGACUCCAAAUUCGAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU_ACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAUCCAC ..............(((.......((((((..((((......((.(....)))....))))..)).)))).......((((((((((.....)))))))))).))).............. (-14.78 = -19.38 + 4.60)

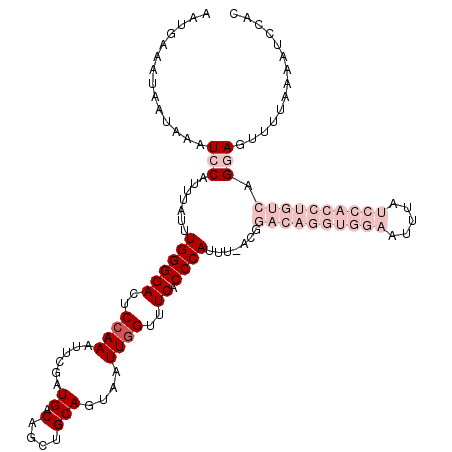
| Location | 2,611,341 – 2,611,455 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -17.52 |
| Energy contribution | -20.28 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2611341 114 - 23771897 GUGGAUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUAAAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUGGAGUCCCCAAAUAAAUGGAUUU--CAUUU---- ..((((((((((......((((((((((.....)))))))))).((((..(((......))).))))................))))))))))(((......)))....--.....---- ( -33.60) >DroSec_CAF1 9525 106 - 1 GUGGAUUUUAAAACUGCUGACAGGUGGAUAAAU--------------AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUGGAGUCCCCAAAUAAAUGGAUUUAUUAUUUAACU ..((((((((((..((.((((((.((.(....(--------------((.(((......))).))).).)).)))))).))..))))))))))(((......)))............... ( -25.70) >DroSim_CAF1 12210 106 - 1 GUGGAUUUUAAAACUGCUGACAGGUGGAUAAAU--------------AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUGGAGUCCCCAAAUAAAUGGAUUUAUUAUUUAACU ..((((((((((..((.((((((.((.(....(--------------((.(((......))).))).).)).)))))).))..))))))))))(((......)))............... ( -25.70) >DroEre_CAF1 9736 116 - 1 ----UUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUUGAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUGGGGUCCCCAAAUAUGAGGAUUUAUUAUUUCAUU ----..........((((((((((((((.....))))))))))(((....((((.((..((((.....................))))..))))))...))))))).............. ( -35.00) >DroYak_CAF1 13002 120 - 1 GCGGAUUUCAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUUGAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUUGGGUCCCCAAAUACAUGGAUUUAUUAUUUCAUU ((((..(((((.......((((((((((.....))))))))))..)))))(((......)))....))))..............(((((....)))))...(((((........))))). ( -34.50) >consensus GUGGAUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGU_AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUCGAAUUUGGAGUCCCCAAAUAAAUGGAUUUAUUAUUUAACU ..((((((((((.....(((((((((((.....)))))............(((......)))..........)))))).....))))))))))(((......)))............... (-17.52 = -20.28 + 2.76)

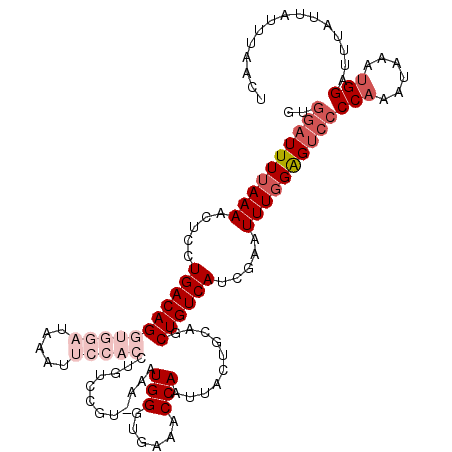
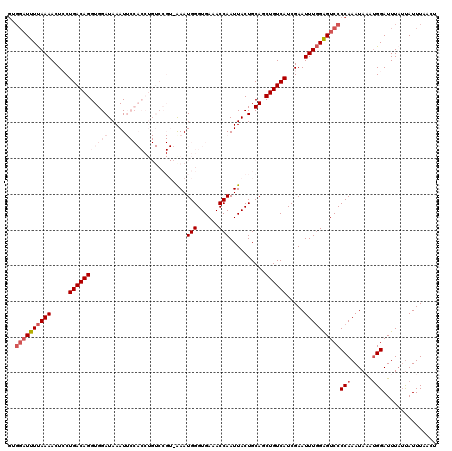
| Location | 2,611,375 – 2,611,485 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -12.92 |
| Energy contribution | -17.00 |
| Covariance contribution | 4.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2611375 110 + 23771897 GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUUUACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAUCCACCUCUUUGCUGCAACAUUUUCUUUCGGCUUA .......(.((((((((..(((...............((((((((((.....)))))))))).(((.........))))))...)))))))).)................ ( -34.90) >DroSec_CAF1 9565 96 + 1 GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU--------------AUUUAUCCACCUGUCAGCAGUUUUAAAAUCCACCUCUUUGCUGGAACAUUUUCUUUCGGCUUA ..((((((.((.(((((.(((......))).))--------------)))....)).)))))).......................(((((((.......)))))))... ( -18.60) >DroSim_CAF1 12250 96 + 1 GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU--------------AUUUAUCCACCUGUCAGCAGUUUUAAAAUCCACCUCUUUGCUGGAGCAUUUUCUUUCGGCUUA ..((((((.((.(((((.(((......))).))--------------)))....)).)))))).......................(((((((.......)))))))... ( -18.30) >DroEre_CAF1 9776 98 + 1 GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUCAACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAA------------UGAAACAUUUUCUUUCGGGCCG ..(((.((((.........)))))))(((..((....((((((((((.....))))))))))..))((((((....------------))))))..........)))... ( -28.10) >DroYak_CAF1 13042 110 + 1 GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUCAACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUGAAAUCCGCCGCUUGGCUGGAGCAUUUUCCUUCGGCUUG ......(((((.((.((...((((((.(((..(....((((((((((.....)))))))))).(((.........)))...)..))).))))))...)).)).))))).. ( -38.50) >consensus GAUGACAGCUGCAGUAAUUGGUUUCACCCAUUU_ACGGACAGGUGGAAUUUAUCCACCUGUCAGGAGUUUUAAAAUCCACCUCUUUGCUGGAACAUUUUCUUUCGGCUUA ......((((.(......(((......))).......((((((((((.....)))))))))).).)))).................(((((((.......)))))))... (-12.92 = -17.00 + 4.08)

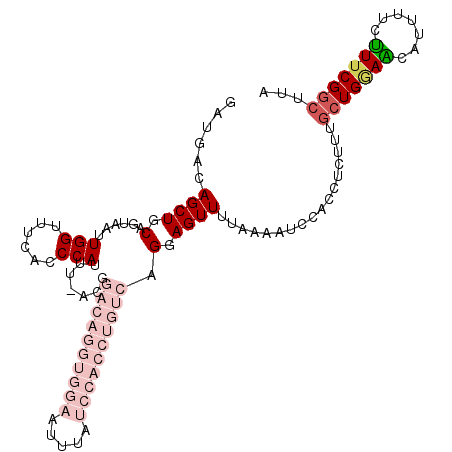
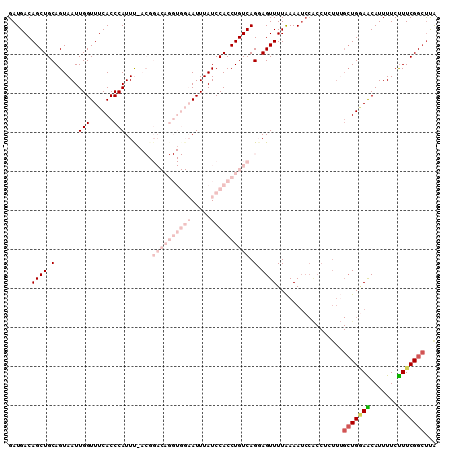
| Location | 2,611,375 – 2,611,485 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -12.76 |
| Energy contribution | -16.72 |
| Covariance contribution | 3.96 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2611375 110 - 23771897 UAAGCCGAAAGAAAAUGUUGCAGCAAAGAGGUGGAUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUAAAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ..........((....(((((((.....(((.(..........).)))((((((((((.....)))))))))).......(((......)))....)))))))..))... ( -34.70) >DroSec_CAF1 9565 96 - 1 UAAGCCGAAAGAAAAUGUUCCAGCAAAGAGGUGGAUUUUAAAACUGCUGACAGGUGGAUAAAU--------------AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ..((((....).......((((.(.....).))))..........)))(((((.((.(....(--------------((.(((......))).))).).)).)))))... ( -21.00) >DroSim_CAF1 12250 96 - 1 UAAGCCGAAAGAAAAUGCUCCAGCAAAGAGGUGGAUUUUAAAACUGCUGACAGGUGGAUAAAU--------------AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ..((((....).......((((.(.....).))))..........)))(((((.((.(....(--------------((.(((......))).))).).)).)))))... ( -20.90) >DroEre_CAF1 9776 98 - 1 CGGCCCGAAAGAAAAUGUUUCA------------UUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUUGAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ((((.(....).....((((((------------(((....(((....((((((((((.....)))))))))).)))....)))))))))...........))))..... ( -32.10) >DroYak_CAF1 13042 110 - 1 CAAGCCGAAGGAAAAUGCUCCAGCCAAGCGGCGGAUUUCAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGUUGAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ......((.(((......))).....(((.((((..(((((.......((((((((((.....))))))))))..)))))(((......)))....)))).))).))... ( -35.20) >consensus UAAGCCGAAAGAAAAUGUUCCAGCAAAGAGGUGGAUUUUAAAACUCCUGACAGGUGGAUAAAUUCCACCUGUCCGU_AAAUGGGUGAAACCAAUUACUGCAGCUGUCAUC ..........((....(((.(((.........................((((((((((.....)))))))))).......(((......)))....))).)))..))... (-12.76 = -16.72 + 3.96)

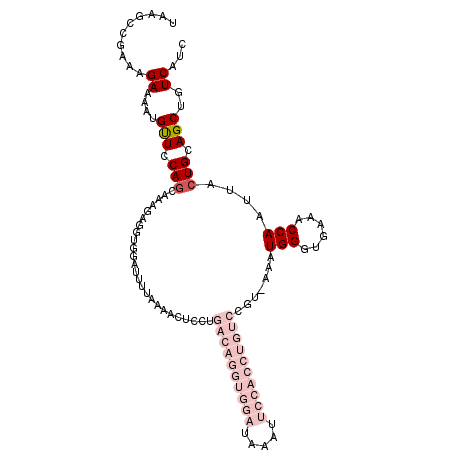
| Location | 2,611,485 – 2,611,597 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -16.78 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2611485 112 + 23771897 UAUAAAUUAAUCAAGCAAUUUAAUGCCAUUUUUUCCACCUCAUGAGCGUUGAAUUAUCCAUCAUUGAGAGUGGAAAAUCCAUU--------UGCAUUUUUUUUUUUUUUUUAGAUUUAAA ..((((((.............(((((....(((((((((((((((..((......))...))).)))).))))))))......--------.)))))...............)))))).. ( -18.99) >DroSec_CAF1 9661 113 + 1 UAUAAUUUAAUCAAGCAAUUUAAUUCCCUUUUUUCCACCCCAAAAACGCUGAAUUAUCCAUCAUUCAGUGUGGAAAAUCCUUCACUAUGAAUGCAUUCU-------UUUUUAGAUUUAAA ...(((((((..(((.(((...((((....((((((((.........(((((((........)))))))))))))))...........))))..)))))-------)..))))))).... ( -16.66) >DroSim_CAF1 12346 114 + 1 UAUAAUUUAAUCAAGCAAUUUAAUUCCCUUUUUUCCACCCCAAAAACGCUGAAUUAUCCAUCAUUCAGAGUGGAAAAUCCUUCACUAUGAAUGCAUUUU------UUUUUUAGAUUUAAA ..............(((.............((((((((..........((((((........)))))).))))))))...(((.....)))))).....------............... ( -14.70) >consensus UAUAAUUUAAUCAAGCAAUUUAAUUCCCUUUUUUCCACCCCAAAAACGCUGAAUUAUCCAUCAUUCAGAGUGGAAAAUCCUUCACUAUGAAUGCAUUUU______UUUUUUAGAUUUAAA ..............(((.............((((((((..........((((((........)))))).))))))))..............))).......................... (-11.45 = -11.56 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:11 2006