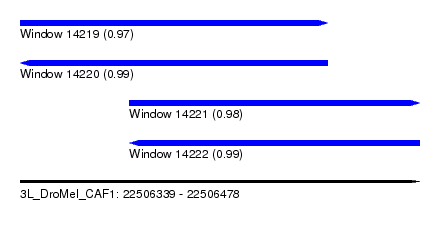
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,506,339 – 22,506,478 |
| Length | 139 |
| Max. P | 0.993480 |

| Location | 22,506,339 – 22,506,446 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.07 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -12.94 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
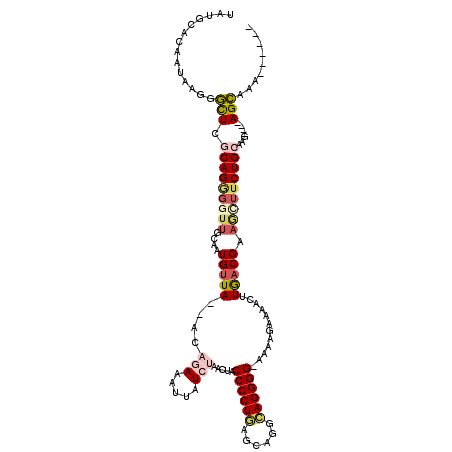
>3L_DroMel_CAF1 22506339 107 + 23771897 UAUGCGCAAUAAGUGCUCGCAGGGGUUGCAAUGUUA--ACAGAAAUUAUCUAACUACCCCUGAGGAGGCAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA------ .............(((((((((..(((((((.(((.--..(((.....))).....((((((......))))))-.......)))))).)))..)..))))..)----))))..------ ( -28.90) >DroVir_CAF1 20534 108 + 1 UAAGCC--------GUUUACAGAUAUCCUUAUGUUA--UUUAACAUUAUCUAACCGCCCCUGUCACGGAAGGGGUAGUUUAUAACUUAACGAUACUUCUGCAAAGU--AGCAGAGAUAAG .....(--------(((...(((((.....(((((.--...))))))))))(((..(((((........)))))..)))........))))....((((((.....--.))))))..... ( -25.40) >DroSec_CAF1 14403 107 + 1 UAUGCACAAUAAGGGCUCUCAGGGGUUGCAAUGUUA--ACAGAAUUUAUCUAACUACCCCUGAGCAGACAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA------ ..............((((((((((((......((((--............)))).))))))))(((((((((..-.........))))........)))))..)----)))...------ ( -27.00) >DroSim_CAF1 14755 107 + 1 UAUGCACAAUAAGGGCUCGCAGGGGUUGCAAUGUUA--ACAGAAUUUAUCUAACUACCCCUGAGCAGGCAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA------ ..............((((((((..(((((((.(((.--..(((.....))).....((((((......))))))-.......)))))).)))..)..))))..)----)))...------ ( -28.00) >DroEre_CAF1 16531 111 + 1 UGUGCAUAAUAGGGGUUCGCAGGGGUUGCAAUGCUA--ACAGAAUUUAUCUGACUACCCCUGCGCAGGCAGGGG-AAGAGAAAACUUGACGAAGCUUCUGCAAGUUACAGCAAA------ ((((.....((((((((((((((..((.........--.((((.....)))).((.(((((((....)))))))-.))..))..)))).)).)))))))).....)))).....------ ( -34.40) >DroMoj_CAF1 27429 110 + 1 CAUGCC--------CCUUGCAGCGUCUGUUAUGUUAUGUUUAACAUUAUCUAGUUACCCCUUAUACAGAAGGGGUGGCAAAAAACUUGACGAAACUCCUGCAAAGC--AGGAGGCGGCUG ...(((--------....((..((((.(((((((((....))))))......((((((((((......))))))))))....)))..))))...(((((((...))--)))))))))).. ( -41.50) >consensus UAUGCACAAUAAGGGCUCGCAGGGGUUGCAAUGUUA__ACAGAAAUUAUCUAACUACCCCUGAGCAGGCAGGGG_AAAAGAAAACUUGACGAAGCUUCUGCAAG____AGCAAA______ ..............(((.(((((((((....(((((....(((.....))).....((((((......))))))............))))).))))))))).......)))......... (-12.94 = -14.12 + 1.17)


| Location | 22,506,339 – 22,506,446 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
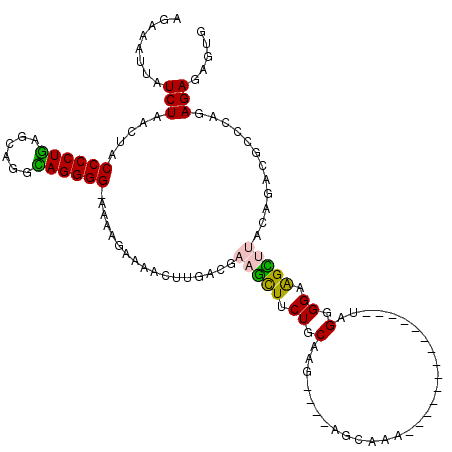
>3L_DroMel_CAF1 22506339 107 - 23771897 ------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGCCUCCUCAGGGGUAGUUAGAUAAUUUCUGU--UAACAUUGCAACCCCUGCGAGCACUUAUUGCGCAUA ------..((((----(..((((((((((....))))..(((...-((((((......)))))).....)))....))))))--.......(((.....))))))))............. ( -25.90) >DroVir_CAF1 20534 108 - 1 CUUAUCUCUGCU--ACUUUGCAGAAGUAUCGUUAAGUUAUAAACUACCCCUUCCGUGACAGGGGCGGUUAGAUAAUGUUAAA--UAACAUAAGGAUAUCUGUAAAC--------GGCUUA .........(((--..((((((((..((((.(((.(((((.(((..(((((........)))))..))).(((...)))..)--)))).))).)))))))))))).--------)))... ( -28.10) >DroSec_CAF1 14403 107 - 1 ------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGUCUGCUCAGGGGUAGUUAGAUAAAUUCUGU--UAACAUUGCAACCCCUGAGAGCCCUUAUUGUGCAUA ------...(((----(..(((((......(..(((....)))..-).....)))))((((((((.((((((.....)))).--.......)).)))))))))))).............. ( -27.60) >DroSim_CAF1 14755 107 - 1 ------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGCCUGCUCAGGGGUAGUUAGAUAAAUUCUGU--UAACAUUGCAACCCCUGCGAGCCCUUAUUGUGCAUA ------...(((----(..((((((((((....))))..(((...-((((((......)))))).....)))....))))))--.......(((.....))))))).............. ( -24.50) >DroEre_CAF1 16531 111 - 1 ------UUUGCUGUAACUUGCAGAAGCUUCGUCAAGUUUUCUCUU-CCCCUGCCUGCGCAGGGGUAGUCAGAUAAAUUCUGU--UAGCAUUGCAACCCCUGCGAACCCCUAUUAUGCACA ------..(((((.((((((..((....))..))))))....((.-(((((((....))))))).)).((((.....)))).--)))))(((((.....)))))................ ( -29.10) >DroMoj_CAF1 27429 110 - 1 CAGCCGCCUCCU--GCUUUGCAGGAGUUUCGUCAAGUUUUUUGCCACCCCUUCUGUAUAAGGGGUAACUAGAUAAUGUUAAACAUAACAUAACAGACGCUGCAAGG--------GGCAUG ..(((.((((((--((...))))))((..((((............(((((((......))))))).........((((((....))))))....))))..))..))--------)))... ( -36.80) >consensus ______UUUGCU____CUUGCAGAAACUUCGUCAAGUUUUCUUUU_CCCCUGCCUGCUCAGGGGUAGUUAGAUAAAUUCUGU__UAACAUUGCAACCCCUGCGAGCCCUUAUUGUGCAUA ................(((((((.......................((((((......))))))....((((.....)))).................)))))))............... (-17.86 = -17.28 + -0.58)



| Location | 22,506,377 – 22,506,478 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.93 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -13.45 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
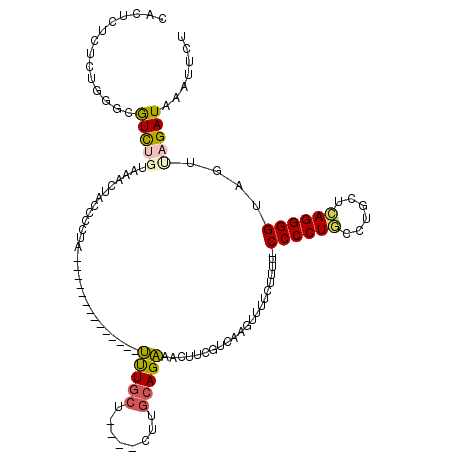
>3L_DroMel_CAF1 22506377 101 + 23771897 AGAAAUUAUCUAACUACCCCUGAGGAGGCAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA--------------UAGGGGUAGCUUACAGACGCCCAGAGAGAGUG ....(((.(((..(((((((((......))))))-...(((((.(((....))))))))((...----.))...--------------)))((((..(.....)..))))..))).))). ( -23.90) >DroGri_CAF1 19160 110 + 1 UAACAUUAUCUAACCACCCCUGAUGCAGUAGGGGUAAUCAGAAACUUGACGAAACUUCUGCAAAGA--GUCAAAAUUGAGAGUA-CAGGAGGGGUAGUA----AAUGA---GAGAGAGAG ...((((..(((.((.((((((.(((.((((((((..((((....))))....)))))))).....--.(((....)))..)))-)))).)))))))..----)))).---......... ( -26.80) >DroSec_CAF1 14441 101 + 1 AGAAUUUAUCUAACUACCCCUGAGCAGACAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA--------------UAGGGGAAGCUUACAGACGUCCAGAGAGAGUG ........(((.....((((((......))))))-.........((.((((((((((((.(...----......--------------..).)))))))).....)))).)))))..... ( -25.00) >DroSim_CAF1 14793 101 + 1 AGAAUUUAUCUAACUACCCCUGAGCAGGCAGGGG-AAAAGAAAACUUGACGAAGUUUCUGCAAG----AGCAAA--------------UAGGGGAAGCUUACAGACGUCCAGAGAGAGUG ........(((.....((((((......))))))-.........((.((((((((((((.(...----......--------------..).)))))))).....)))).)))))..... ( -25.20) >DroEre_CAF1 16569 105 + 1 AGAAUUUAUCUGACUACCCCUGCGCAGGCAGGGG-AAGAGAAAACUUGACGAAGCUUCUGCAAGUUACAGCAAA--------------UAGGGGUAGUUUGCAGACGCGCAGAGAGAGUG ...((((.((((.((.(((((((....)))))))-.))...............((.(((((((..(((..(...--------------..)..)))..))))))).)).))))..)))). ( -37.40) >DroMoj_CAF1 27461 111 + 1 UAACAUUAUCUAGUUACCCCUUAUACAGAAGGGGUGGCAAAAAACUUGACGAAACUCCUGCAAAGC--AGGAGGCGGCUGGCUAUUAUGAGGGGAGGUC----AAUGC---GAGAGAGAG ........(((.((((((((((......))))))))))......((((.((...(((((((...))--)))))..((((..((........))..))))----..)))---))))))... ( -36.70) >consensus AGAAAUUAUCUAACUACCCCUGAGCAGGCAGGGG_AAAAGAAAACUUGACGAAGCUUCUGCAAG____AGCAAA______________UAGGGGAAGCUUACAGACGCCCAGAGAGAGUG ........(((.....((((((......)))))).................(((((.((.(.............................).)).)))))............)))..... (-13.45 = -12.87 + -0.58)


| Location | 22,506,377 – 22,506,478 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.93 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22506377 101 - 23771897 CACUCUCUCUGGGCGUCUGUAAGCUACCCCUA--------------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGCCUCCUCAGGGGUAGUUAGAUAAUUUCU ..(((.....))).(((((...(((((((((.--------------..(((.----...)))(((((((....))))))).....-.............))))))))))))))....... ( -25.20) >DroGri_CAF1 19160 110 - 1 CUCUCUCUC---UCAUU----UACUACCCCUCCUG-UACUCUCAAUUUUGAC--UCUUUGCAGAAGUUUCGUCAAGUUUCUGAUUACCCCUACUGCAUCAGGGGUGGUUAGAUAAUGUUA .........---..(((----((((((((((....-.....(((....))).--....(((((.((....((((......)))).....)).)))))..)))))))).)))))....... ( -23.50) >DroSec_CAF1 14441 101 - 1 CACUCUCUCUGGACGUCUGUAAGCUUCCCCUA--------------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGUCUGCUCAGGGGUAGUUAGAUAAAUUCU ...(((..((((((((((((((((........--------------...)))----..)))))).....))))............-((((((......)))))))))..)))........ ( -25.20) >DroSim_CAF1 14793 101 - 1 CACUCUCUCUGGACGUCUGUAAGCUUCCCCUA--------------UUUGCU----CUUGCAGAAACUUCGUCAAGUUUUCUUUU-CCCCUGCCUGCUCAGGGGUAGUUAGAUAAAUUCU ...(((..((((((((((((((((........--------------...)))----..)))))).....))))............-((((((......)))))))))..)))........ ( -24.20) >DroEre_CAF1 16569 105 - 1 CACUCUCUCUGCGCGUCUGCAAACUACCCCUA--------------UUUGCUGUAACUUGCAGAAGCUUCGUCAAGUUUUCUCUU-CCCCUGCCUGCGCAGGGGUAGUCAGAUAAAUUCU ...(((..((((((.(((((((..(((..(..--------------...)..)))..))))))).))..................-.((((((....))))))))))..)))........ ( -30.40) >DroMoj_CAF1 27461 111 - 1 CUCUCUCUC---GCAUU----GACCUCCCCUCAUAAUAGCCAGCCGCCUCCU--GCUUUGCAGGAGUUUCGUCAAGUUUUUUGCCACCCCUUCUGUAUAAGGGGUAACUAGAUAAUGUUA .........---(((((----(((..............((.....))(((((--((...)))))))....)))))......))).(((((((......)))))))............... ( -25.80) >consensus CACUCUCUCUGGGCGUCUGUAAACUACCCCUA______________UUUGCU____CUUGCAGAAACUUCGUCAAGUUUUCUUUU_CCCCUGCCUGCUCAGGGGUAGUUAGAUAAAUUCU ..............(((((...........................(((((........)))))......................((((((......))))))....)))))....... (-14.35 = -14.35 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:05 2006