
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,505,270 – 22,505,403 |
| Length | 133 |
| Max. P | 0.989357 |

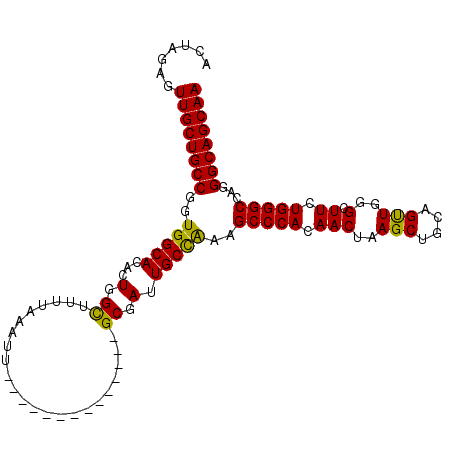
| Location | 22,505,270 – 22,505,364 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.12 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22505270 94 + 23771897 ACUAUAAUUGCUGCCGGUGGCACACUGGUUUUAAAAUU--------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGGUUCUGGGCCAGGGCAGCAA .......((((((((..((((.((.(.((.........--------------)).).))(((.(((((.(((((.......)))))))))).))))))).)))))))) ( -35.30) >DroSec_CAF1 13346 94 + 1 ACUAGAGUUGCUGCCGGUGGCACACUGGCUUUUAAAUU--------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAA .......((((((((..((((.((.(.((.........--------------)).).))((((((((((..(((.......)))))))))).))))))).)))))))) ( -35.20) >DroSim_CAF1 13701 94 + 1 ACUAGAGUUGCUGCCGGUGGCACACUGGCUUUUAAAUU--------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAA .......((((((((..((((.((.(.((.........--------------)).).))((((((((((..(((.......)))))))))).))))))).)))))))) ( -35.20) >DroEre_CAF1 15417 108 + 1 ACUAGAUUUGCUGCCGCUGGCACACUGGCUUUCAAAUUGCGAAUUGCGAAUUGCGAUUGCUGCAGCCCACAACUUAGCUGCAGCUGGGUUUCUGGGCCGGGGCAGCAA .......((((((((.(((((.((..(((((...((((((((........))))))))((((((((..........)))))))).)))))..)).))))))))))))) ( -47.90) >consensus ACUAGAGUUGCUGCCGGUGGCACACUGGCUUUUAAAUU______________GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAA .......((((((((..(((((...(.((.......................)).).)))))..(((((.(((..(((....)))..).)).)))))...)))))))) (-29.88 = -29.12 + -0.75)

| Location | 22,505,270 – 22,505,364 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -28.89 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22505270 94 - 23771897 UUGCUGCCCUGGCCCAGAACCCCAACUGCAGCUUAGUUGUGGGCUUUGGCAAUCGC--------------AAUUUUAAAACCAGUGUGCCACCGGCAGCAAUUAUAGU ((((((((.(((((((((.(((((((((.....)))))).))).)))))....(((--------------.............))).))))..))))))))....... ( -34.32) >DroSec_CAF1 13346 94 - 1 UUGCUGCCCUGGCCCAGAAGCCCAACUGCAGCUUAGUUGUGGGCUUUGGCAAUCGC--------------AAUUUAAAAGCCAGUGUGCCACCGGCAGCAACUCUAGU ((((((((.((((.((((((((((...((......))..))))))))(((......--------------.........)))..)).))))..))))))))....... ( -36.86) >DroSim_CAF1 13701 94 - 1 UUGCUGCCCUGGCCCAGAAGCCCAACUGCAGCUUAGUUGUGGGCUUUGGCAAUCGC--------------AAUUUAAAAGCCAGUGUGCCACCGGCAGCAACUCUAGU ((((((((.((((.((((((((((...((......))..))))))))(((......--------------.........)))..)).))))..))))))))....... ( -36.86) >DroEre_CAF1 15417 108 - 1 UUGCUGCCCCGGCCCAGAAACCCAGCUGCAGCUAAGUUGUGGGCUGCAGCAAUCGCAAUUCGCAAUUCGCAAUUUGAAAGCCAGUGUGCCAGCGGCAGCAAAUCUAGU (((((((((.(((.((........(((((((((........)))))))))..(((.((((.((.....))))))))).......)).))).).))))))))....... ( -40.90) >consensus UUGCUGCCCUGGCCCAGAAGCCCAACUGCAGCUUAGUUGUGGGCUUUGGCAAUCGC______________AAUUUAAAAGCCAGUGUGCCACCGGCAGCAACUCUAGU ((((((((..(((.((((((((((...((......))..)))))))).((....))............................)).)))...))))))))....... (-28.89 = -29.45 + 0.56)


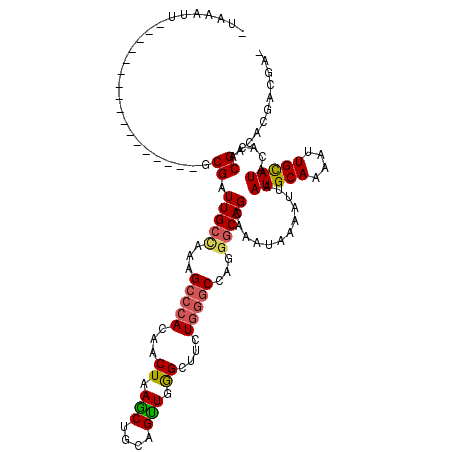
| Location | 22,505,302 – 22,505,403 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22505302 101 + 23771897 -AAAAUU-----------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGGUUCUGGGCCAGGGCAGCAAAUAAAAUUAAUGCAAAAUUGCAUACACACGACACGACGA- -....((-----------------((...((((...(((((.((((.(((....)))..)))).)))))...)))))))).........(((((....)))))................- ( -28.00) >DroSec_CAF1 13378 101 + 1 -UAAAUU-----------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAAAUAAAAUUAAUGCAAAAUUGCAUACACACGACACGACGA- -.....(-----------------((((((......((((.(((((.......)))))((((....)))).)))).(((...........)))..))))))).................- ( -25.70) >DroSim_CAF1 13733 101 + 1 -UAAAUU-----------------GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAAAUAAAAUUAAUGCAAAAUUGCAUACACACGACACGACGA- -.....(-----------------((((((......((((.(((((.......)))))((((....)))).)))).(((...........)))..))))))).................- ( -25.70) >DroEre_CAF1 15449 115 + 1 -CAAAUUGCGAAUUGCGAAUU---GCGAUUGCUGCAGCCCACAACUUAGCUGCAGCUGGGUUUCUGGGCCGGGGCAGCAAAUAAAAUUAAUGCAAAAUUGCAUACACACGACCCGACGA- -.....(((((.((((.((((---....(((((((.(((((.((((((((....))))))))..)))))....)))))))....))))...))))..))))).................- ( -37.80) >DroMoj_CAF1 26407 110 + 1 UUGGACGGC--AUUUCGAAGCACGUCGUUUGAAAAUG--UA-CGCUAAACUGCAGUUAAGA--CUUGGCC---GCAGCAACAAAAAUUAAUGCAAAAUUGUAUACACACGACACGAAUAU .....((((--........))..(((((.((......--..-.......((((.((((...--..)))).---))))............(((((....))))).)).))))).))..... ( -19.50) >consensus _UAAAUU_________________GCGAUUGCCAAAGCCCACAACUAAGCUGCAGUUGGGCUUCUGGGCCAGGGCAGCAAAUAAAAUUAAUGCAAAAUUGCAUACACACGACACGACGA_ .........................((.(((((...(((((...((.(((....))).))....)))))...)))))............(((((....))))).....)).......... (-17.22 = -17.46 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:00 2006