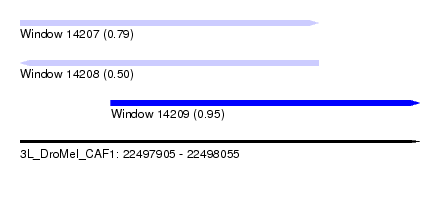
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,497,905 – 22,498,055 |
| Length | 150 |
| Max. P | 0.947161 |

| Location | 22,497,905 – 22,498,017 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -35.46 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22497905 112 + 23771897 GCUCCAGCACUUUCAGCUGCUUUUGAUUGCGUGAAGCAGCUUUCUCUUCAGCUUUCGGGCUUUGUUUUGUCGCCGGCUGAGUUUUUGGCCAGACGGAGCAGGGACUGAGAAG (((..((.......(((((((((.........)))))))))....))..)))((((((.((((((((((((...(((..(....)..))).)))))))))))).)))))).. ( -39.80) >DroSec_CAF1 6320 112 + 1 GCUCCAGCACUUUCAGCUGCUUUUGAUUGCGUGAAGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGACUGAGCGACUGAGGAG ((....))......(((((((((.........)))))))))..((((((((((....))))((((((.(((((.((((((....)))))).).)))).))))))..)))))) ( -40.10) >DroSim_CAF1 6345 112 + 1 GCUCCAGCACUUUCAGCUGCUUUUGAUUGCGUGAAGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGACUGAGCGACCGAGGAG ((....))......(((((((((.........)))))))))..((((((((((....))).((((((.(((((.((((((....)))))).).)))).)))))).))))))) ( -41.60) >DroEre_CAF1 8388 112 + 1 GCCCCAGCACUUUCAGCUGCUUUUGAUUGCGUGAAGCAGCUUUCUCUUCGGCUUUCAGGCUUUGUUGUGUCGCCGGCUGAGUUUUUAGCCAGACGGAGCAGCGACUGAGCAG (((..((.......(((((((((.........)))))))))....))..))).(((((.(.(((((.((((...((((((....)))))).)))).))))).).)))))... ( -36.60) >consensus GCUCCAGCACUUUCAGCUGCUUUUGAUUGCGUGAAGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGAAGCAGCGACUGAGGAG ((....))......(((((((((.........)))))))))..((((((((((....))).((((((((((((.((((((....)))))).).))))))))))).))))))) (-35.46 = -36.40 + 0.94)

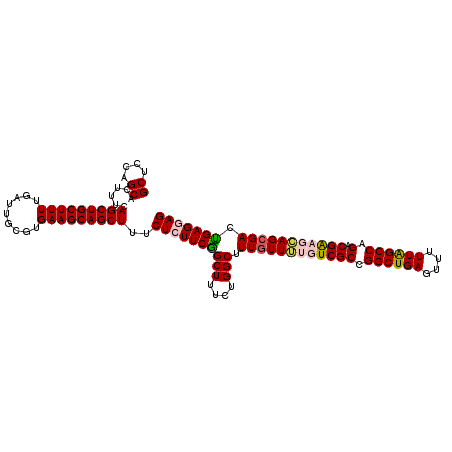
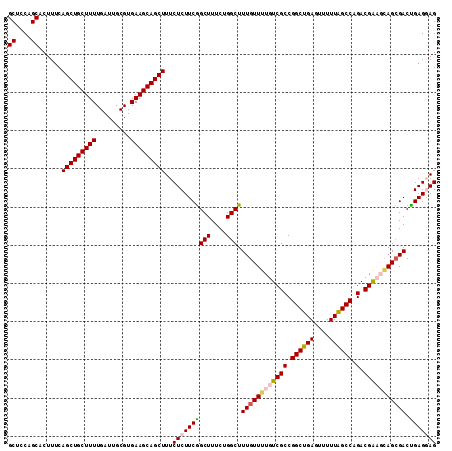
| Location | 22,497,905 – 22,498,017 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -26.44 |
| Energy contribution | -27.75 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22497905 112 - 23771897 CUUCUCAGUCCCUGCUCCGUCUGGCCAAAAACUCAGCCGGCGACAAAACAAAGCCCGAAAGCUGAAGAGAAAGCUGCUUCACGCAAUCAAAAGCAGCUGAAAGUGCUGGAGC .............(((((....((((......(((((.(((...........))).....)))))......((((((((...........))))))))....).)))))))) ( -28.30) >DroSec_CAF1 6320 112 - 1 CUCCUCAGUCGCUCAGUCGUCUGGCUAAAAACUCAGCCGGCGACAAAACAAAGCCAGAAAGCCGAAGAGAAAGCUGCUUCACGCAAUCAAAAGCAGCUGAAAGUGCUGGAGC ((((.((.((.(((.((((.((((((........))))))))))........((......))....)))..((((((((...........))))))))))...))..)))). ( -34.10) >DroSim_CAF1 6345 112 - 1 CUCCUCGGUCGCUCAGUCGUCUGGCUAAAAACUCAGCCGGCGACAAAACAAAGCCAGAAAGCCGAAGAGAAAGCUGCUUCACGCAAUCAAAAGCAGCUGAAAGUGCUGGAGC ((((.(..((.(((.((((.((((((........))))))))))........((......))....)))..((((((((...........))))))))))..)....)))). ( -34.80) >DroEre_CAF1 8388 112 - 1 CUGCUCAGUCGCUGCUCCGUCUGGCUAAAAACUCAGCCGGCGACACAACAAAGCCUGAAAGCCGAAGAGAAAGCUGCUUCACGCAAUCAAAAGCAGCUGAAAGUGCUGGGGC ...(((((.((((.(((((.((((((........))))))))..........((......))....)))..((((((((...........))))))))...))))))))).. ( -33.60) >consensus CUCCUCAGUCGCUCAGCCGUCUGGCUAAAAACUCAGCCGGCGACAAAACAAAGCCAGAAAGCCGAAGAGAAAGCUGCUUCACGCAAUCAAAAGCAGCUGAAAGUGCUGGAGC ....((((.((((..((((.((((((........))))))))))...........................((((((((...........))))))))...))))))))... (-26.44 = -27.75 + 1.31)

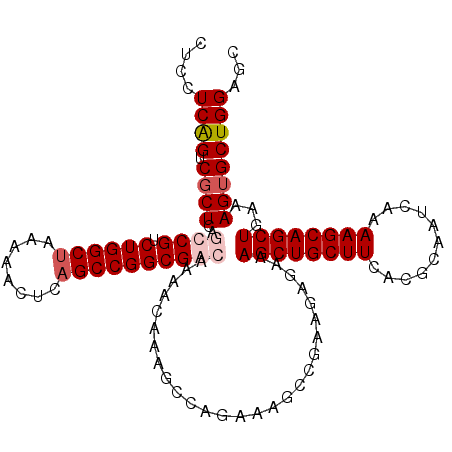
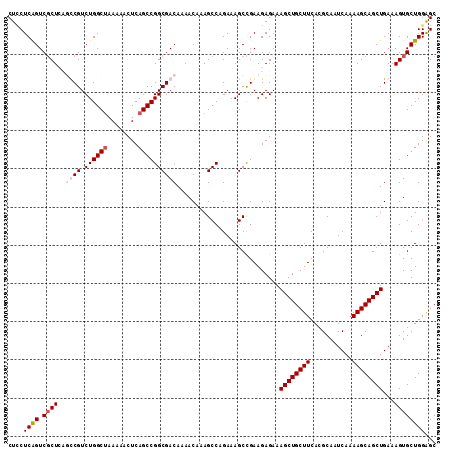
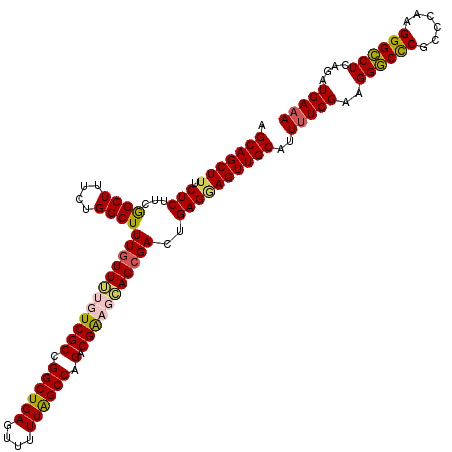
| Location | 22,497,939 – 22,498,055 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -41.69 |
| Energy contribution | -42.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22497939 116 + 23771897 AGCAGCUUUCUCUUCAGCUUUCGGGCUUUGUUUUGUCGCCGGCUGAGUUUUUGGCCAGACGGAGCAGGGACUGAGAAGUUGCAUU-UCGAAGGGCCCGCCCG-GGGCCUCAAAUGAAA .(((((((.(((.((((((....))))((((((((((...(((..(....)..))).)))))))))).))..)))))))))).((-(((..((((((.....-))))))....))))) ( -45.80) >DroSec_CAF1 6354 118 + 1 AGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGACUGAGCGACUGAGGAGUUGCAUUUUCGAAGGGCCCACCCAAGGGCCUCAGAUGAAA .(((((((.(((...((((....))))((((((.(((((.((((((....)))))).).)))).))))))..))))))))))..(((((..((((((......))))))....))))) ( -45.50) >DroSim_CAF1 6379 118 + 1 AGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGACUGAGCGACCGAGGAGUUGCAUUUUCGAAGGGCCCGCCCAAGGGCCUCAGAUGAAA .(((((((.(((...((((....))))((((((.(((((.((((((....)))))).).)))).))))))..))))))))))..(((((..((((((......))))))....))))) ( -46.50) >DroEre_CAF1 8422 118 + 1 AGCAGCUUUCUCUUCGGCUUUCAGGCUUUGUUGUGUCGCCGGCUGAGUUUUUAGCCAGACGGAGCAGCGACUGAGCAGUUGCAUUUUCGAAGGACUCGCCCGAGGGUCUCGGAUGAAA .......((((((..(((((((.(((..((((.((((...((((((....)))))).)))).))))((((((....))))))....((....))...))).)))))))..))).))). ( -41.70) >consensus AGCAGCUUUCUCUUCGGCUUUCUGGCUUUGUUUUGUCGCCGGCUGAGUUUUUAGCCAGACGAAGCAGCGACUGAGGAGUUGCAUUUUCGAAGGGCCCGCCCAAGGGCCUCAGAUGAAA .(((((((.(((...((((....))))((((((((((((.((((((....)))))).).)))))))))))..))))))))))..(((((..((((((......))))))....))))) (-41.69 = -42.12 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:53 2006