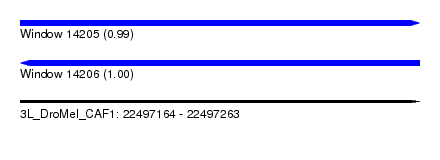
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,497,164 – 22,497,263 |
| Length | 99 |
| Max. P | 0.998404 |

| Location | 22,497,164 – 22,497,263 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
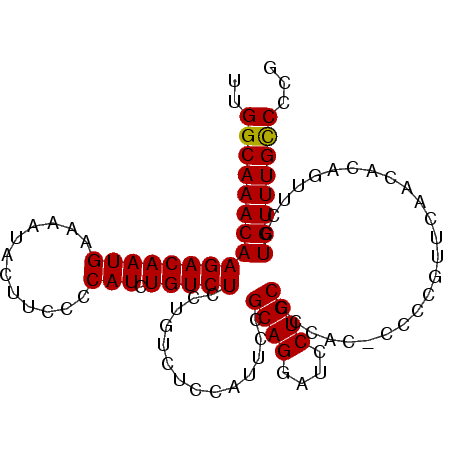
| Reading direction | forward |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22497164 99 + 23771897 CGGGGCAAACAGGAUCU-UGUUAAAGGGGG-GUAGGGCAGGAUCCUGCGGAAAGGAGACAGGAGACAGAUGGGGAAGUAUUUUCAUUGUCUUGUUUGCCAA ...((((((((((((((-((((........-....)))))))))))........(((((.(....).((((..((....))..)))))))))))))))).. ( -37.00) >DroSec_CAF1 5567 101 + 1 CGGGGCAAACAGGAACUGCGUUGAACGGGGGGUGGGGCAGGAUCCUGCGGAAUGGAGAAAGGAGACAGAUGGGAAAGUAUUUUCAUUGUCUUGUUUGCCAA ...(((((((((((.((.((((...((.(((((........))))).)).)))).))...(....).((((..((....))..)))).))))))))))).. ( -35.90) >DroSim_CAF1 5589 100 + 1 CGGGGCAAACAGGAACUGUGUUGAACGGGGGGUG-GGCAGGAUCCUGCGGAAUGGAGACCGGAGACAGAUGGGGAAGUAUUUUCAUUGUCUUGUUUGCCAA ...(((((((((((.((((.((...((.(((((.-......))))).))....(....)..)).))))(((..((....))..)))..))))))))))).. ( -34.90) >DroEre_CAF1 7648 97 + 1 CGGGACAAACAGGAAUUGCGUUG---GGGG-AUGGGGCAGGAUCCUGCGGAAGGGGAACAGGAGACAGAUGGGGAAGUAUUUUCAUUGUCUUGUUUGCCAA .........(((((.((((.(((---....-.))).))))..))))).....((.((((((((....((((..((....))..)))).)))))))).)).. ( -29.10) >consensus CGGGGCAAACAGGAACUGCGUUGAACGGGG_GUGGGGCAGGAUCCUGCGGAAUGGAGACAGGAGACAGAUGGGGAAGUAUUUUCAUUGUCUUGUUUGCCAA ...(((((((((((......................((((....))))............(....).((((..((....))..)))).))))))))))).. (-28.14 = -28.20 + 0.06)

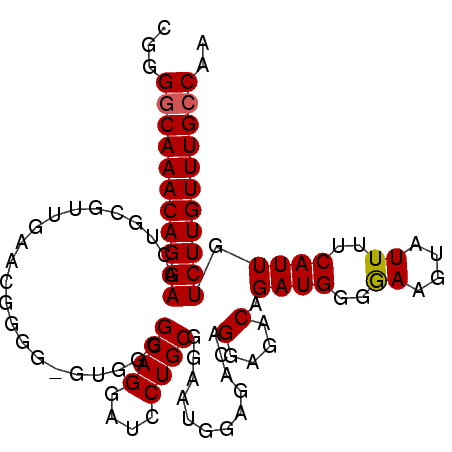
| Location | 22,497,164 – 22,497,263 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -20.81 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22497164 99 - 23771897 UUGGCAAACAAGACAAUGAAAAUACUUCCCCAUCUGUCUCCUGUCUCCUUUCCGCAGGAUCCUGCCCUAC-CCCCCUUUAACA-AGAUCCUGUUUGCCCCG ..((((((((((((((((............))).)))))..))).........((((((((.((......-..........))-.)))))))))))))... ( -21.59) >DroSec_CAF1 5567 101 - 1 UUGGCAAACAAGACAAUGAAAAUACUUUCCCAUCUGUCUCCUUUCUCCAUUCCGCAGGAUCCUGCCCCACCCCCCGUUCAACGCAGUUCCUGUUUGCCCCG ..(((((((.((((((((..((....))..))).)))))................((((..((((.................)))).)))))))))))... ( -21.93) >DroSim_CAF1 5589 100 - 1 UUGGCAAACAAGACAAUGAAAAUACUUCCCCAUCUGUCUCCGGUCUCCAUUCCGCAGGAUCCUGCC-CACCCCCCGUUCAACACAGUUCCUGUUUGCCCCG ..((((((((((((((((............))).))))).(((..........((((....)))).-......)))..............))))))))... ( -22.23) >DroEre_CAF1 7648 97 - 1 UUGGCAAACAAGACAAUGAAAAUACUUCCCCAUCUGUCUCCUGUUCCCCUUCCGCAGGAUCCUGCCCCAU-CCCC---CAACGCAAUUCCUGUUUGUCCCG ..((((((((((((((((............))).)))))..............((((....)))).....-....---............))))))))... ( -18.60) >consensus UUGGCAAACAAGACAAUGAAAAUACUUCCCCAUCUGUCUCCUGUCUCCAUUCCGCAGGAUCCUGCCCCAC_CCCCGUUCAACACAGUUCCUGUUUGCCCCG ..((((((((((((((((............))).)))))..............((((....)))).........................))))))))... (-20.81 = -20.62 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:50 2006