
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,483,215 – 22,483,360 |
| Length | 145 |
| Max. P | 0.834166 |

| Location | 22,483,215 – 22,483,330 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.38 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -9.26 |
| Energy contribution | -9.59 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
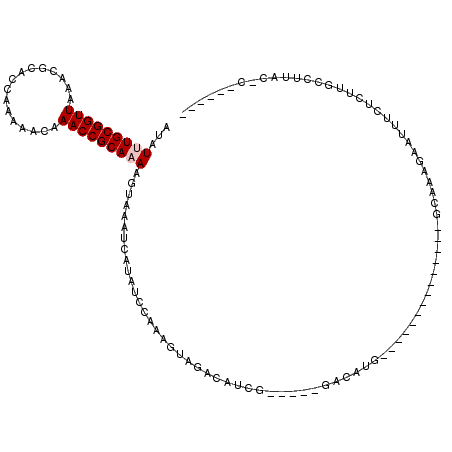
>3L_DroMel_CAF1 22483215 115 + 23771897 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCAAAAGUAAAUCAUAUCCAGAGCAGGCAUUG-----GACAUGGCCAAGCAUCCGAGGCAAAGAAUUUCUCUUGCCUCACCCGACCGU ...(((((((((................)))))))))......((((.(((((.(....).)))-----)).))))..........(((((((((.....)).))))))).......... ( -29.49) >DroVir_CAF1 680 91 + 1 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCAAAAGUAAAUCAUACCCAAAGUAGACACCG-----AGCAUG--------------GCCAAGAAUUUCUCUGUGCUU---------- ...(((((((((................)))))))))..........................(-----((((((--------------(...((.....))))))))))---------- ( -16.99) >DroPse_CAF1 13968 95 + 1 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCACAAGUAAAUCAUAUCCAGAGUAGACAUCG-----GACAUG--------------CCAAGGAAUUUCCUUUGCUCUACUC------ .....(((((((................))))))).................(((((((((..(-----((.((.--------------(....).)).)))..)).)))))))------ ( -21.49) >DroYak_CAF1 769 115 + 1 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCAAAAGUAAAUCAUAUCCAAAACAGACAUUG-----GACACGGCCAAGCAUCCAAGGCAAGGAAUUUCUCUUGCCUCACCCGACCGU ...(((((((((................)))))))))...........(((((........)))-----)).((((...........((((((((......)))))))).......)))) ( -26.26) >DroMoj_CAF1 545 96 + 1 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCAAAAGUAAAUCAUACCCAAAGUAGACACCGGACCGGACAUG--------------GCCAAGAAUUUCUCUCGACUU---------- ...(((((((((................)))))))))...............((((.((....((.(((....))--------------)))..((.....)))).))))---------- ( -15.59) >DroPer_CAF1 13944 95 + 1 AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCACAAGUAAAUCAUAUCCAGAGUAGACAUCG-----GACAUG--------------CCAAGGAAUUUCCUUUGCUCUACUC------ .....(((((((................))))))).................(((((((((..(-----((.((.--------------(....).)).)))..)).)))))))------ ( -21.49) >consensus AUAUUUGCGGUUAAACGCACCAAAAACAAACCGCAAAAGUAAAUCAUAUCCAAAGUAGACAUCG_____GACAUG______________GCAAAGAAUUUCUCUUGCCUUAC_C______ ...(((((((((................)))))))))................................................................................... ( -9.26 = -9.59 + 0.33)

| Location | 22,483,215 – 22,483,330 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.38 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
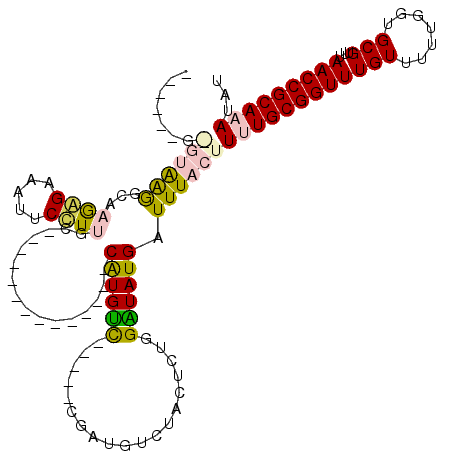
>3L_DroMel_CAF1 22483215 115 - 23771897 ACGGUCGGGUGAGGCAAGAGAAAUUCUUUGCCUCGGAUGCUUGGCCAUGUC-----CAAUGCCUGCUCUGGAUAUGAUUUACUUUUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ..((((((((((((((.(((.....)))))))))....))))))))(((((-----((..(....)..)))))))........((((((((((((.......)))...)))))))))... ( -40.30) >DroVir_CAF1 680 91 - 1 ----------AAGCACAGAGAAAUUCUUGGC--------------CAUGCU-----CGGUGUCUACUUUGGGUAUGAUUUACUUUUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ----------..((...(((.....))).))--------------((((((-----(((........))))))))).......((((((((((((.......)))...)))))))))... ( -23.20) >DroPse_CAF1 13968 95 - 1 ------GAGUAGAGCAAAGGAAAUUCCUUGG--------------CAUGUC-----CGAUGUCUACUCUGGAUAUGAUUUACUUGUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ------(((((((((.((((.....)))).)--------------)(((((-----((..(....)..)))))))..))))))).((((((((((.......)))...)))))))..... ( -27.80) >DroYak_CAF1 769 115 - 1 ACGGUCGGGUGAGGCAAGAGAAAUUCCUUGCCUUGGAUGCUUGGCCGUGUC-----CAAUGUCUGUUUUGGAUAUGAUUUACUUUUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ((((((((((((((((((.(.....)))))))))....))))))))))(((-----(((........))))))..........((((((((((((.......)))...)))))))))... ( -42.20) >DroMoj_CAF1 545 96 - 1 ----------AAGUCGAGAGAAAUUCUUGGC--------------CAUGUCCGGUCCGGUGUCUACUUUGGGUAUGAUUUACUUUUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ----------..((((((((...))))))))--------------(((..((((...(((....)))))))..))).......((((((((((((.......)))...)))))))))... ( -24.30) >DroPer_CAF1 13944 95 - 1 ------GAGUAGAGCAAAGGAAAUUCCUUGG--------------CAUGUC-----CGAUGUCUACUCUGGAUAUGAUUUACUUGUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU ------(((((((((.((((.....)))).)--------------)(((((-----((..(....)..)))))))..))))))).((((((((((.......)))...)))))))..... ( -27.80) >consensus ______G_GUAAGGCAAGAGAAAUUCCUUGC______________CAUGUC_____CGAUGUCUACUCUGGAUAUGAUUUACUUUUGCGGUUUGUUUUUGGUGCGUUUAACCGCAAAUAU .......((((((...((((.....))))................((((((...................)))))).))))))((((((((((((.......)))...)))))))))... (-14.31 = -14.18 + -0.13)

| Location | 22,483,255 – 22,483,360 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.23 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -26.27 |
| Energy contribution | -27.50 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22483255 105 - 23771897 UCUAAUAUUUUUGCCUACA-----AAUGCCGGACGACGGUCGGGUGAGGCAAGAGAAAUUCUUUGCCUCGGAUGCUUGGCCAUGUCCAAUGCCUGCUCUGGAUAUGAUUU ............(((....-----...(((.(((....))).)))((((((.(((.....)))))))))........)))((((((((..(....)..)))))))).... ( -34.60) >DroEre_CAF1 844 108 - 1 UCUAAU-UUUUUGCCUACGAGUUGAGUUCCGGACGACUUUUGGGGGCGGCAAGAGAAAUUCCUUGCCUCGGAUGCUUGGCCACGUCC-AUGUCUGUUUUAGAUAUGAUUU (((((.-......((((.((((((.........)))))).))))(((((((((.(.....)))))))..(((((........)))))-..)))....)))))........ ( -29.30) >DroYak_CAF1 809 105 - 1 UCUAAUAUUUUUGCCUUCA-----AGUUCCGGACGACGGUCGGGUGAGGCAAGAGAAAUUCCUUGCCUUGGAUGCUUGGCCGUGUCCAAUGUCUGUUUUGGAUAUGAUUU ...................-----......((((.((((((((((((((((((.(.....)))))))))....)))))))))))))).((((((.....))))))..... ( -36.80) >consensus UCUAAUAUUUUUGCCUACA_____AGUUCCGGACGACGGUCGGGUGAGGCAAGAGAAAUUCCUUGCCUCGGAUGCUUGGCCAUGUCCAAUGUCUGUUUUGGAUAUGAUUU ............(((............(((.(((....))).)))((((((((.(.....)))))))))........)))(((((((((........))))))))).... (-26.27 = -27.50 + 1.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:47 2006