
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,474,459 – 22,474,593 |
| Length | 134 |
| Max. P | 0.703266 |

| Location | 22,474,459 – 22,474,575 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -14.60 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.98 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
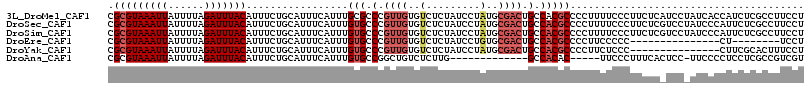
>3L_DroMel_CAF1 22474459 116 - 23771897 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCGCCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCAUCCUAUCACCAUCUCGCCUUCCU .((((((((.....((((........))))......)))))))).(((.(((((.(........).))).)).)))........................................ ( -15.10) >DroSec_CAF1 24570 116 - 1 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCGUCCUAUCCCAUUCUCGCCUUCCU .(((..(((.....((((..((((.....((((.......))))....))))..))))......(((.......))).........................)))..)))...... ( -14.20) >DroSim_CAF1 24573 116 - 1 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCCCUUUUCCCUUCUCGUCCUAUCCCAUUCUCGCCUUCCU .(((..(((.....((((..((((.....((((.......))))....))))..))))......(((.......))).........................)))..)))...... ( -14.20) >DroEre_CAF1 28244 93 - 1 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAUCCUGUGCGACUGCCACGCCCCUUCCCCC---------------CU--------UCCU .(((((((((......))))))).................(((.(.((((((.(........))))))).).)))))..........---------------..--------.... ( -14.10) >DroYak_CAF1 30045 101 - 1 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCCCUUCUCCC---------------CUUCGCACUUUCCU .(((((((((......)))))))......((((.......)))).(((.(((((.(........).))).)).)))...........---------------....))........ ( -15.10) >DroAna_CAF1 22251 97 - 1 CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCGGCUGUCUCUUG-------------GCCACAC-----UUCCCUUUCACUCC-UUCCCCUCCUCGCCGUCGU .(((((((((......)))))))......((((.......)))).(((((.....))-------------)))....-----..............-...........))...... ( -14.90) >consensus CGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUCUCUAUCCUAUGCGACUGCCACGCCCCUUUUCCCUUCUC_UCC_AUCCCCUUCUCGCCUUCCU .(((((((((......))))))).................(((.(.((((..(.........)..)))).).)))))....................................... ( -8.93 = -9.98 + 1.06)

| Location | 22,474,496 – 22,474,593 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -10.02 |
| Energy contribution | -9.93 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22474496 97 - 23771897 ----------AACUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCGCCCGUUGUGUC-UCUAUCCUAUGCGACUGCCACGCCC ----------....((((((.((((.((.((((((((.....((((........))))......)))))))))).))))((.-..........))))))))....... ( -24.10) >DroVir_CAF1 69163 99 - 1 CAACGGCAACAACUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCUCAG--CUCGCACACUUAGCCAU-------CCACAUUC ....(((......(((.(((......)))(((((((((......)))))))......(((.......)))...)--)..)))......)))..-------........ ( -19.70) >DroGri_CAF1 26728 104 - 1 CAACAACGGCAACUGCAGUCAACAAAGAAGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCUCAG--UGCGUCCACUUGGCCAC--CACGCCCACGCCC .......(((.((((..........(((((.(((((((......))))))).)))))(((.......))).)))--)((((((....))....--.))))....))). ( -21.90) >DroEre_CAF1 28258 97 - 1 ----------AACUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUC-UCUAUCCUGUGCGACUGCCACGCCC ----------....((((((.(((.(((((((((((((......)))))))......((((.......)))).....)))))-)......)))..))))))....... ( -24.30) >DroYak_CAF1 30067 97 - 1 ----------AACUGCAGUCAACAAAGGCGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCCGUUGUGUC-UCUAUCCUAUGCGACUGCCACGCCC ----------....((((((.....(((((((((((((......)))))))......((((.......)))).....)))))-)(........).))))))....... ( -21.70) >DroAna_CAF1 22284 82 - 1 ----------AACUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGUGCCGGCUGUCUC-UUG-------------GCCACAC-- ----------....((.(((......)))))(((((((......)))))))......((((.......)))).(((((....-.))-------------)))....-- ( -17.80) >consensus __________AACUGCAGUCAACAAAGACGCGUAAAUUAUUUUAGAUUUACAUUUCUGCAUUUCAUUUGCGCCCGUUGUGUC_UCUAUCCUAU__GACUGCCACGCCC .............(((((((......)))..(((((((......))))))).....))))................................................ (-10.02 = -9.93 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:43 2006