| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,472,821 – 22,472,924 |
| Length | 103 |
| Max. P | 0.919837 |

| Location | 22,472,821 – 22,472,924 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -18.29 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22472821 103 + 23771897 UGUGUGAGUGCGCGGUUAGCAA----------AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGC------UGCAGGCUAUGAAAAAUGCAGCUGAAGAGAUCUC ..((.((((((((((((.....----------))))))....)))))).))...((((......)))).((....((((------((((............))))))))...))..... ( -28.60) >DroVir_CAF1 66918 96 + 1 ---AUGUGUGUGUGGUUGGAAA----------AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGCCAAAGCUGCA-GCUAUGAAAAAUGUAUUCUG--------- ---..((((.((..(((.....----------)))..))...))))(((((...((((......))))(((...(((((....))))))-))..)))))...........--------- ( -23.60) >DroGri_CAF1 24797 97 + 1 U--GUGUAUAUGUGGCUAGGAA----------AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGCCAGAGUUGCA-GCUAUGAAAAAUGCAGUCAG--------- .--((((((.((..(((.....----------)))..)).))))))(((((...((((......))))(((...(((((....))))))-))..)))))...........--------- ( -26.10) >DroEre_CAF1 26607 113 + 1 UGUGUAAGUGCGCGGUUAGCAAAUGCUGCAAAAGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGC------UGCAGGCCAUGAAAAAUGCAGCUGGAGAGAGCUC .((((....)))).....(((..(((.((....)).)))..)))(((((((((.((((......)))))))....((((------((((............))))))))..)))))).. ( -36.00) >DroYak_CAF1 28305 103 + 1 UGUGCGAGUGCGCGGUUAGCAA----------AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGC------UGCAGGCUAUGAAAAAUUCAGCUGGAGAGAGCUU ...((.(((((((((((.....----------))))))....)))))(((((((((............((....))(((------.....))).........)))))))))....)).. ( -27.10) >DroAna_CAF1 20675 102 + 1 -AUGUAUGUGCGCGGUUGGAAA----------AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCUGC------UGCAGGCUAUGAAAAAUGCAUCCGAGAAAACCUG -......((((((((((.....----------))))))......((((.((((.((((......))))((....)).))------)).))))..........))))............. ( -22.20) >consensus UGUGUGAGUGCGCGGUUAGCAA__________AGCUGCAAAUGCGCUUUCAGUUGAUUAAAUUAAAUCGCUAAAGCAGC______UGCAGGCUAUGAAAAAUGCAGCCGGAGAGA_CU_ ...............(((((............(((((((...(((((((.(((.((((......)))))))))))).))......)))).)))............)))))......... (-18.29 = -17.60 + -0.69)


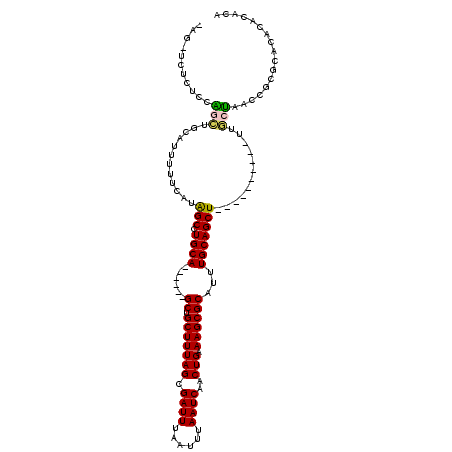
| Location | 22,472,821 – 22,472,924 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -16.11 |
| Energy contribution | -15.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22472821 103 - 23771897 GAGAUCUCUUCAGCUGCAUUUUUCAUAGCCUGCA------GCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU----------UUGCUAACCGCGCACUCACACA (((.......((((((((............))))------))))..(((((((((......)))).....(((((((....)).)))----------))))))).......)))..... ( -27.50) >DroVir_CAF1 66918 96 - 1 ---------CAGAAUACAUUUUUCAUAGC-UGCAGCUUUGGCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU----------UUUCCAACCACACACACAU--- ---------(((...............((-((((((....)))))...)))((((......))))..)))(((((((....)).)))----------)).................--- ( -20.50) >DroGri_CAF1 24797 97 - 1 ---------CUGACUGCAUUUUUCAUAGC-UGCAACUCUGGCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU----------UUCCUAGCCACAUAUACAC--A ---------.....((((.((.....)).-))))....(((((((....))((((......))))....((((((((....)).)))----------)))..))))).........--. ( -20.20) >DroEre_CAF1 26607 113 - 1 GAGCUCUCUCCAGCUGCAUUUUUCAUGGCCUGCA------GCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCUUUUGCAGCAUUUGCUAACCGCGCACUUACACA ..(((.....((((((((............))))------))))....)))((((......))))........((((.......((....))(((....)))....))))......... ( -28.90) >DroYak_CAF1 28305 103 - 1 AAGCUCUCUCCAGCUGAAUUUUUCAUAGCCUGCA------GCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU----------UUGCUAACCGCGCACUCGCACA ..((......((((((................))------))))..(((((((((......)))).....(((((((....)).)))----------)))))))..))((....))... ( -23.19) >DroAna_CAF1 20675 102 - 1 CAGGUUUUCUCGGAUGCAUUUUUCAUAGCCUGCA------GCAGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU----------UUUCCAACCGCGCACAUACAU- ((((((......((........))..))))))..------((.((......((((......))))....((((((((....)).)))----------)))......))))........- ( -18.30) >consensus _AG_UCUCUCCAGCUGCAUUUUUCAUAGCCUGCA______GCUGCUUUAGCGAUUUAAUUUAAUCAACUGAAAGCGCAUUUGCAGCU__________UUGCUAACCGCGCACACACACA ...........(((............(((.((((......((.(((((((.((((......))))..))).))))))...)))))))............)))................. (-16.11 = -15.67 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:41 2006