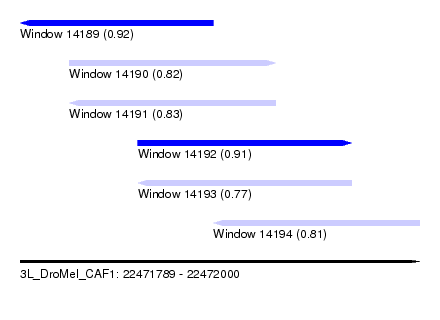
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,471,789 – 22,472,000 |
| Length | 211 |
| Max. P | 0.916283 |

| Location | 22,471,789 – 22,471,891 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -29.17 |
| Energy contribution | -28.58 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471789 102 - 23771897 GCUGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUGGGCGGGAGAAACUUCUGUUGUAGAGCGCUAAAGUUGGCCCAAGGCUAGUGAAAAACGAGAA- (((((((((((.....((((((.(.....)...)))))))))))(((...(((((..((........))..)))))..)))..))).)))............- ( -37.20) >DroSec_CAF1 21932 103 - 1 ACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUUAGCCCAAGGCUAGUAAAAAACGAGAAA (((((((((.......((((((.(.....)...))))))..)))((((..(((((..((.((...))))..))))).))))..)))).))............. ( -35.00) >DroSim_CAF1 21917 103 - 1 ACAGCCGCCCAUUGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUUAGCCCAAGGCUAGUAAAAAACGAGAAA (((((((((.......((((((.(.....)...))))))..)))((((..(((((..((.((...))))..))))).))))..)))).))............. ( -35.00) >DroEre_CAF1 25625 103 - 1 GCAGCCGCCCACGGCUGCUGGCGGAUUUUCAUAGCCGGCUCGGCGGGCGAAACUUCCCUUGCAGAGCGGCAAAGUUGCCCCAAGGCUGGUGAAAAACGAAACA (((((((....)))))))...((..(((((((((((.((((.(((((.((....)).))))).))))((((....))))....)))).))))))).))..... ( -45.00) >DroYak_CAF1 27490 84 - 1 -------------------CGCGGAUUUUCAUAGCCAGCUCGGCGGACGAAACUUCUGUCGCAGAGCGCUAAAGUUGGCCCAAGGCUAGUAAAAAACGAAAUA -------------------..((..((((..(((((.((((.((((..((....))..)))).))))((((....))))....)))))..))))..))..... ( -25.80) >consensus ACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUUGGCCCAAGGCUAGUAAAAAACGAGAAA .((((.((.....)).)))).((..((((..(((((.((((.((((..((....))..)))).))))((((....))))....)))))..))))..))..... (-29.17 = -28.58 + -0.59)

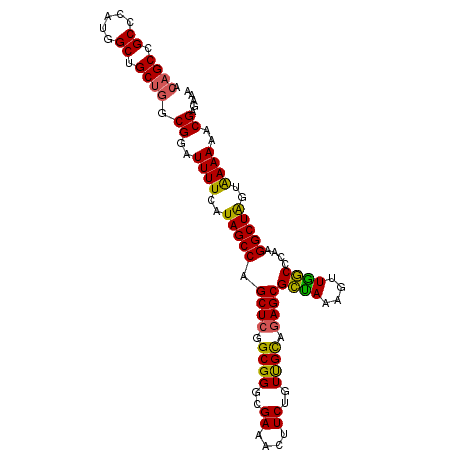

| Location | 22,471,815 – 22,471,924 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -50.61 |
| Consensus MFE | -36.90 |
| Energy contribution | -38.40 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471815 109 + 23771897 AACUUUAGCGCUCUACAACAGAAGUUUCUCCCGCCCAGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCAGCUUUGGUUGGGCGUGGCUGCAGCGAGCUGUC-------CAC .......((((((.((....((((((....(((((((((((((.((....))..)))))).....))))))).)))))).)).))))))((..((((....)))))-------).. ( -43.30) >DroSec_CAF1 21959 109 + 1 AACUUUAGCGCUCUGCAACAGAAGUUUCGCCCGCCGAGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCUGUUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG-------CGC .......((((.((((......(((..((((((((..((((((.((....))..))))))((((......))))......))))))))..))).......)))).)-------))) ( -52.62) >DroSim_CAF1 21944 109 + 1 AACUUUAGCGCUCUGCAACAGAAGUUUCGCCCGCCGAGCUGGCUAUGAAAAUCCGCCAGCAGCAAUGGGCGGCUGUUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG-------CGC .......((((.((((......(((..((((((((..((((((.((....))..))))))(((........)))......))))))))..))).......)))).)-------))) ( -49.02) >DroEre_CAF1 25652 116 + 1 AACUUUGCCGCUCUGCAAGGGAAGUUUCGCCCGCCGAGCCGGCUAUGAAAAUCCGCCAGCAGCCGUGGGCGGCUGCUUUGGGUGGGCGUGGCUGCAGCGAGCAGCGCGCAGCGCGC ......(((((((.((..(((........))))).))).)))).........(((((((((((((....))))))))...)))))(((((.((((.(((.....)))))))))))) ( -57.50) >consensus AACUUUAGCGCUCUGCAACAGAAGUUUCGCCCGCCGAGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCUGCUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG_______CGC .........((((.((......(((..((((((((..((((((...........))))))((((......))))......))))))))..)))...)))))).............. (-36.90 = -38.40 + 1.50)

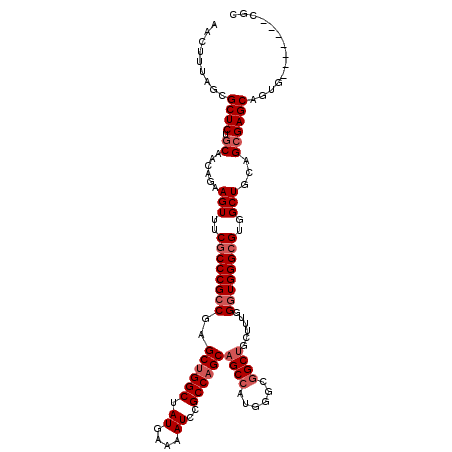

| Location | 22,471,815 – 22,471,924 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.19 |
| Mean single sequence MFE | -46.35 |
| Consensus MFE | -35.21 |
| Energy contribution | -36.15 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471815 109 - 23771897 GUG-------GACAGCUCGCUGCAGCCACGCCCAACCAAAGCUGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUGGGCGGGAGAAACUUCUGUUGUAGAGCGCUAAAGUU ...-------...(((.(.(((((((..((((((.........((((....)))).((((((.(.....)...))))))))))))((((....))))))))))).).)))...... ( -41.10) >DroSec_CAF1 21959 109 - 1 GCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUU (((-------(.((((.....((((...(((((.((......(((((....)))))((((((.(.....)...))))))..)).)))))......)))).)))).))))....... ( -46.40) >DroSim_CAF1 21944 109 - 1 GCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUUGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUU (((-------(.((((.....((((...(((((.((......(((........)))((((((.(.....)...))))))..)).)))))......)))).)))).))))....... ( -42.40) >DroEre_CAF1 25652 116 - 1 GCGCGCUGCGCGCUGCUCGCUGCAGCCACGCCCACCCAAAGCAGCCGCCCACGGCUGCUGGCGGAUUUUCAUAGCCGGCUCGGCGGGCGAAACUUCCCUUGCAGAGCGGCAAAGUU ((((...))))(((((((..(((((((.((((...((..((((((((....))))))))(((.(.....)...)))))...)))))))((....))...)))))))))))...... ( -55.50) >consensus GCG_______CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCUCGGCGGGCGAAACUUCUGUUGCAGAGCGCUAAAGUU ..............((((((.((((...(((((.((......(((((....)))))((((((.(.....)...))))))..)).)))))......)))).)).))))......... (-35.21 = -36.15 + 0.94)


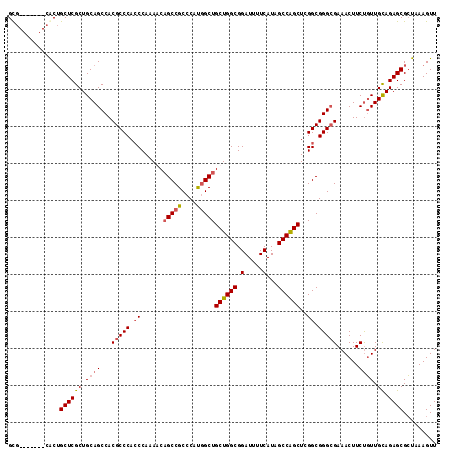
| Location | 22,471,851 – 22,471,964 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -46.88 |
| Consensus MFE | -38.70 |
| Energy contribution | -39.08 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471851 113 + 23771897 AGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCAGCUUUGGUUGGGCGUGGCUGCAGCGAGCUGUC-------CACAGCGAGACGAGCUUGCAGUUUUUAAAGAACUGCUAUUUUU ((((.(((.........(((..(((((((((..((((.......))))..))))))))).))).((((..-------..)))).)).).)))).(((((((.....)))))))....... ( -43.30) >DroSec_CAF1 21995 113 + 1 AGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCUGUUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG-------CGCAGCGAGACGAGCGUGCAGUUUUUAAAGAACUGCUAUUUUU .(((.(((........(((((.((((((......))))))....)))))(((((.((((.....))))))-------)))....)).).)))..(((((((.....)))))))....... ( -44.70) >DroSim_CAF1 21980 113 + 1 AGCUGGCUAUGAAAAUCCGCCAGCAGCAAUGGGCGGCUGUUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG-------CGCAGCGAGACGAGCGUGCAGUUUUUAAAGAACUGCUAUUUUU .(((.(((........(((((.(((((........)))))....)))))(((((.((((.....))))))-------)))....)).).)))..(((((((.....)))))))....... ( -41.10) >DroEre_CAF1 25688 120 + 1 AGCCGGCUAUGAAAAUCCGCCAGCAGCCGUGGGCGGCUGCUUUGGGUGGGCGUGGCUGCAGCGAGCAGCGCGCAGCGCGCAGCGAGAUGAGCGUGCAGUUUUUAGAGAGCUGCUAUUUUU (((.((((.((((((((((((((((((((....))))))))...)))))(((((.((((.....))))))))).((((((..........)))))).)))))))...)))))))...... ( -58.40) >consensus AGCUGGCUAUGAAAAUCCGCCAGCAGCCAUGGGCGGCUGCUUUGGGUGGGCGUGGCUGCAGCGAGCAGUG_______CGCAGCGAGACGAGCGUGCAGUUUUUAAAGAACUGCUAUUUUU .((.(((((((....(((((((((((((......)))))))...))))))))))))))).((..((.((((.....)))).)).......))..(((((((.....)))))))....... (-38.70 = -39.08 + 0.37)



| Location | 22,471,851 – 22,471,964 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.79 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471851 113 - 23771897 AAAAAUAGCAGUUCUUUAAAAACUGCAAGCUCGUCUCGCUGUG-------GACAGCUCGCUGCAGCCACGCCCAACCAAAGCUGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCU .......((((((.......))))))......(.((.((((((-------((.((..(((((((((((.((.((........))..))...))))))).))))..)))))))))).))). ( -36.80) >DroSec_CAF1 21995 113 - 1 AAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCU .......((((((.......))))))..(..(((...((((((-------(.......)).))))).)))..).........(((((....)))))((((((.(.....)...)))))). ( -34.40) >DroSim_CAF1 21980 113 - 1 AAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUUGCUGCUGGCGGAUUUUCAUAGCCAGCU .......((((((.......))))))..(..(((...((((((-------(.......)).))))).)))..).........(((........)))((((((.(.....)...)))))). ( -30.40) >DroEre_CAF1 25688 120 - 1 AAAAAUAGCAGCUCUCUAAAAACUGCACGCUCAUCUCGCUGCGCGCUGCGCGCUGCUCGCUGCAGCCACGCCCACCCAAAGCAGCCGCCCACGGCUGCUGGCGGAUUUUCAUAGCCGGCU .......(((((............(((.((.......)))))((((...)))).....)))))((((............((((((((....))))))))(((.(.....)...))))))) ( -44.40) >consensus AAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG_______CACUGCUCGCUGCAGCCACGCCCACCCAAAACAGCCGCCCAUGGCUGCUGGCGGAUUUUCAUAGCCAGCU .......((((((.......))))))...........(((((..........((((.....))))...((((.......((((((((....))))))))))))..........).)))). (-27.85 = -28.79 + 0.94)



| Location | 22,471,891 – 22,472,000 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22471891 109 - 23771897 CCGACAAAGCUUUGCACAAAAAGUAAAACCAACUUCAAAAAUAGCAGUUCUUUAAAAACUGCAAGCUCGUCUCGCUGUG-------GACAGCUCGCUGCAGCCACGCCCAACCAAA ..(((..((((((((.......)))))................((((((.......))))))..))).)))..((.(((-------(.(((....)))...))))))......... ( -23.60) >DroSec_CAF1 22035 98 - 1 CC-----------GCACAAAAAGUAAAACCAACUUCAAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAA ..-----------((.....((((.......))))........((((((.......))))))..)).(((...((((((-------(.......)).))))).))).......... ( -19.20) >DroSim_CAF1 22020 109 - 1 CCGACAAAGCUUCGCACAAAAAGUAAAACCAACUUCAAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG-------CACUGCUCGCUGCAGCCACGCCCACCCAAA ..(((..(((..........((((.......))))........((((((.......))))))..))).)))..((((((-------(.......)).))))).............. ( -21.70) >DroEre_CAF1 25728 116 - 1 CCGGCAAGGCUUGCCACAAAAAGCAGUACAAACUUCAAAAAUAGCAGCUCUCUAAAAACUGCACGCUCAUCUCGCUGCGCGCUGCGCGCUGCUCGCUGCAGCCACGCCCACCCAAA ..(((..(((((((........)))..................(((((............(((.((.......)))))((((...)))).....)))))))))..)))........ ( -30.90) >DroYak_CAF1 27574 93 - 1 CCGGCAAAGCUUGGCACAAAAAGAA-AACCAACUUCAAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGCCUCGCU--------------------UGCAGCCACGCCCAUCCA-- ..(((.((((..(((.......(((-.......))).......((((((.......))))))......)))..)))--------------------)...)))...........-- ( -23.10) >consensus CCGACAAAGCUUGGCACAAAAAGUAAAACCAACUUCAAAAAUAGCAGUUCUUUAAAAACUGCACGCUCGUCUCGCUGCG_______CACUGCUCGCUGCAGCCACGCCCACCCAAA .............((............................((((((.......))))))...........(((((...................)))))...))......... (-10.75 = -11.43 + 0.68)

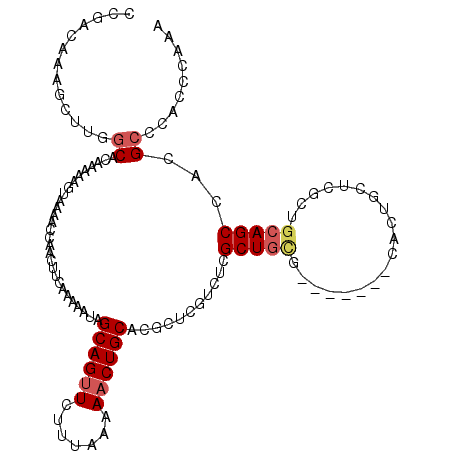
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:39 2006