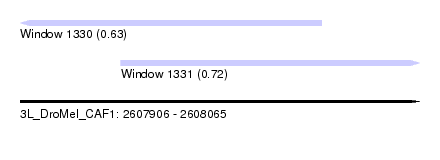
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,607,906 – 2,608,065 |
| Length | 159 |
| Max. P | 0.723772 |

| Location | 2,607,906 – 2,608,026 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2607906 120 - 23771897 AGAAAUUCUGUGUAUUUUGCACAUUAUUUCUAUACAUUUUAUGCAGUUUCUGCAUGAGUUAUGACCAGCUUAUUGAGUACUGUAUAAUUUGAAAGUAUAUGUUUCAUUAAGACCAUAUUU ((((((..(((((.....)))))..))))))...((..((((((((((((...(((((((......))))))).))).)))))))))..)).((((((..((((.....)))).)))))) ( -28.20) >DroSec_CAF1 6325 118 - 1 AGAAACUCUGUGUU-UUUGCAAAUUAUUUCUGUGCAUGUUAUGCAGUUCCUA-AUGAGUUAUGGCCAGCUUAUUGAGUACUGUAUAAUUUGAAAGUAUAUGUUUCAUUAAGACCAUAUUU .(((((..((((((-((((((...........)))..((((((((((.(.((-(((((((......))))))))).).))))))))))..))))))))).)))))............... ( -25.90) >DroSim_CAF1 8817 118 - 1 AGAAACUCUGUGUA-UUUGCACAUUAUUUCUGUACAUUUUAUGCAGUUUCUA-AUGAGUUAUGGCCAGCUUAUUGAGUACUGUACAAUUUGAAAGUAUAUGUUUCAUUAAGACCAUAUUU .(((((..(((((.-...))))).......(((((.((((((((((((((.(-(((((((......))))))))))).)))))).....)))))))))).)))))............... ( -25.50) >DroEre_CAF1 6387 118 - 1 ACAAUCUCUGUGUA-UUUGCAAAUUAUUUCUAUAUAUUUUAUGCAGUUUCUA-AUGAGUUAUGGGCAGCUUAUUGUCUGCUGCAGAAUUUUAAAGUAUAUGUUUCAUUUAGACCACAGUU .......(((((..-................((((((((((((((((...((-(((((((......)))))))))...))))))......))))))))))((((.....))))))))).. ( -24.60) >consensus AGAAACUCUGUGUA_UUUGCAAAUUAUUUCUAUACAUUUUAUGCAGUUUCUA_AUGAGUUAUGGCCAGCUUAUUGAGUACUGUAUAAUUUGAAAGUAUAUGUUUCAUUAAGACCAUAUUU .(((((..(((((.....))))).......(((((...(((((((((......(((((((......))))))).....))))))))).......))))).)))))............... (-17.84 = -18.15 + 0.31)

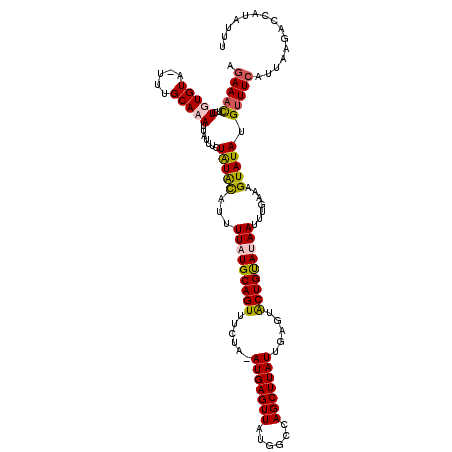
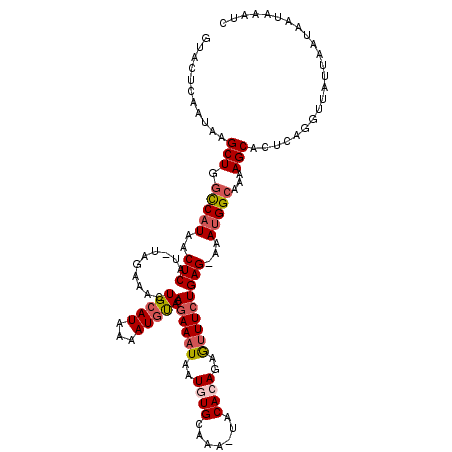
| Location | 2,607,946 – 2,608,065 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.98 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2607946 119 + 23771897 GUACUCAAUAAGCUGGUCAUAACUCAUGCAGAAACUGCAUAAAAUGUAUAGAAAUAAUGUGCAAAAUACACAGAAUUUCUGAG-AAACGGCAAAAGCACUAUGGUUAUUAAUAAUAAAUC ...........(((.(((....(((((((((...)))))).........((((((..((((.......))))..)))))))))-....)))...)))....................... ( -23.00) >DroSec_CAF1 6365 117 + 1 GUACUCAAUAAGCUGGCCAUAACUCAU-UAGGAACUGCAUAACAUGCACAGAAAUAAUUUGCAAA-AACACAGAGUUUCUGAG-AAAUGGCCAAAGCACUCAGGUUAUUAAUAAUAAAUC ...........(((((((((...((.(-((((((((((((...)))).((((.....))))....-.......))))))))))-).))))))..)))....................... ( -24.00) >DroSim_CAF1 8857 117 + 1 GUACUCAAUAAGCUGGCCAUAACUCAU-UAGAAACUGCAUAAAAUGUACAGAAAUAAUGUGCAAA-UACACAGAGUUUCUGAG-AAAUGGCAAAAGCACUCAGGUUAUUAAUAAUAAAUC ...........(((.(((((...((.(-((((((((........((((((.......))))))..-.......))))))))))-).)))))...)))....................... ( -25.73) >DroEre_CAF1 6427 118 + 1 GCAGACAAUAAGCUGCCCAUAACUCAU-UAGAAACUGCAUAAAAUAUAUAGAAAUAAUUUGCAAA-UACACAGAGAUUGUGAGGAAAUGGUGAAAGCCUACUAGUUAAUAAUAAUCAAUC ..........(((((((((((((((..-.......((((....................))))..-......))).))))).))....(((....)))...))))).............. ( -15.98) >consensus GUACUCAAUAAGCUGGCCAUAACUCAU_UAGAAACUGCAUAAAAUGUACAGAAAUAAUGUGCAAA_UACACAGAGUUUCUGAG_AAAUGGCAAAAGCACUCAGGUUAUUAAUAAUAAAUC ...........(((.(((((..(((..........(((((...))))).((((((..((((.......))))..)))))))))...)))))...)))....................... (-14.79 = -15.98 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:06 2006