
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,452,395 – 22,452,506 |
| Length | 111 |
| Max. P | 0.744878 |

| Location | 22,452,395 – 22,452,506 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -13.60 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
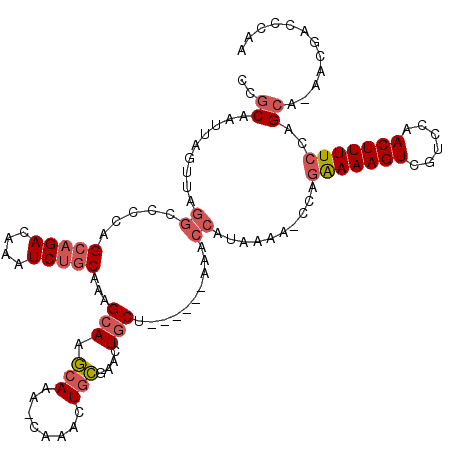
>3L_DroMel_CAF1 22452395 111 + 23771897 CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA-CAAACUGCGAACUGCU------AAACCAUAAAA-CCAGAAAAGUCGUCCAACUUUUCCAGCA-AACGACCCAA ..((.........((.....(((((....)))))..((((.(((..-.....)))....))))------...))......-...(((((((......)))))))..)).-.......... ( -21.10) >DroPse_CAF1 17238 110 + 1 CCACAAUUA----------AGCAGACAAGUCCCCGAAGCAAAUAAAAGAAGCUCUGGAGUCCGUCCUGCAAUCCAUGAAGUCCGUGAAAGUCGGUCAACUUUUUUAGCGCUACGAGCCAA .........----------.((.((....))..((.(((...(((((((.((...(((.....))).))..........(.((((....).))).)...)))))))..))).)).))... ( -17.50) >DroSec_CAF1 3751 111 + 1 CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA-CAAACUGCGAACUGCU------AAACCAUAAAA-CCAGAAAAGUCGUCCAACUUUUCUAGCA-AACGACCCAA ..((.........((.....(((((....)))))..((((.(((..-.....)))....))))------...))......-..((((((((......)))))))).)).-.......... ( -22.70) >DroSim_CAF1 3755 111 + 1 CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA-CAAACUGCGAACUGCU------AAACCAUAAAA-CCAGAAAAGUCGUCCAACUUUUCCAGCA-AACGACCCAA ..((.........((.....(((((....)))))..((((.(((..-.....)))....))))------...))......-...(((((((......)))))))..)).-.......... ( -21.10) >DroEre_CAF1 8062 100 + 1 CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA-CAAACUGCGAAAUGCU------AAACCAUAAAA-CCAGAAAAGUCGUCCAACUUUUCCAGC------------ ..((.........((.....(((((....)))))..((((.(((..-.....)))....))))------...))......-...(((((((......)))))))..))------------ ( -19.40) >DroYak_CAF1 9260 111 + 1 CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA-CAAACUGCGAACUGCU------AAACCAUAAAA-CCAGAAAAGUCGUCCAACUUUUCCAGCA-AACGACCCAA ..((.........((.....(((((....)))))..((((.(((..-.....)))....))))------...))......-...(((((((......)))))))..)).-.......... ( -21.10) >consensus CCGCAAUUAGUUAGGCCCCAGCAGACAAAUCUGCAAAGCAAGCAAA_CAAACUGCGAACUGCU______AAACCAUAAAA_CCAGAAAAGUCGUCCAACUUUUCCAGCA_AACGACCCAA ..((.........((.....(((((....)))))...(((.(((........)))....)))..........))..........(((((((......)))))))..))............ (-13.60 = -14.52 + 0.92)

| Location | 22,452,395 – 22,452,506 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
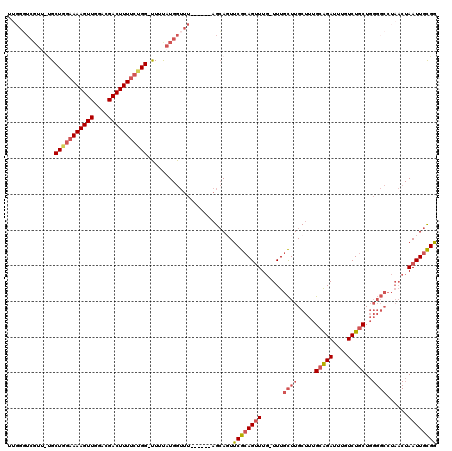
>3L_DroMel_CAF1 22452395 111 - 23771897 UUGGGUCGUU-UGCUGGAAAAGUUGGACGACUUUUCUGG-UUUUAUGGUUU------AGCAGUUCGCAGUUUG-UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG ((((..(((.-.((..((((((((....))))))))..)-)...)))..))------)).....(((((((.(-((.....((((((((((....))))).)))))..))).))))))). ( -37.30) >DroPse_CAF1 17238 110 - 1 UUGGCUCGUAGCGCUAAAAAAGUUGACCGACUUUCACGGACUUCAUGGAUUGCAGGACGGACUCCAGAGCUUCUUUUAUUUGCUUCGGGGACUUGUCUGCU----------UAAUUGUGG (((((.......))))).((((((....))))))(((((.((....))...(((((..((.((((..(((...........)))..)))).))..))))).----------...))))). ( -25.90) >DroSec_CAF1 3751 111 - 1 UUGGGUCGUU-UGCUAGAAAAGUUGGACGACUUUUCUGG-UUUUAUGGUUU------AGCAGUUCGCAGUUUG-UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG ((((..(((.-.((((((((((((....)))))))))))-)...)))..))------)).....(((((((.(-((.....((((((((((....))))).)))))..))).))))))). ( -38.30) >DroSim_CAF1 3755 111 - 1 UUGGGUCGUU-UGCUGGAAAAGUUGGACGACUUUUCUGG-UUUUAUGGUUU------AGCAGUUCGCAGUUUG-UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG ((((..(((.-.((..((((((((....))))))))..)-)...)))..))------)).....(((((((.(-((.....((((((((((....))))).)))))..))).))))))). ( -37.30) >DroEre_CAF1 8062 100 - 1 ------------GCUGGAAAAGUUGGACGACUUUUCUGG-UUUUAUGGUUU------AGCAUUUCGCAGUUUG-UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG ------------((..((((((((....))))))))..)-)..........------.......(((((((.(-((.....((((((((((....))))).)))))..))).))))))). ( -32.20) >DroYak_CAF1 9260 111 - 1 UUGGGUCGUU-UGCUGGAAAAGUUGGACGACUUUUCUGG-UUUUAUGGUUU------AGCAGUUCGCAGUUUG-UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG ((((..(((.-.((..((((((((....))))))))..)-)...)))..))------)).....(((((((.(-((.....((((((((((....))))).)))))..))).))))))). ( -37.30) >consensus UUGGGUCGUU_UGCUGGAAAAGUUGGACGACUUUUCUGG_UUUUAUGGUUU______AGCAGUUCGCAGUUUG_UUUGCUUGCUUUGCAGAUUUGUCUGCUGGGGCCUAACUAAUUGCGG .............(((((((((((....)))))))))))........................((((((((......((((.....(((((....)))))..))))......)))))))) (-22.50 = -23.50 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:25 2006