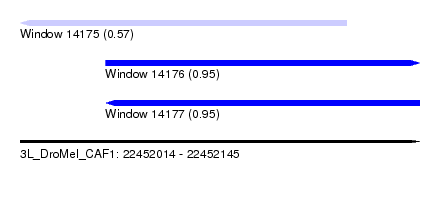
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,452,014 – 22,452,145 |
| Length | 131 |
| Max. P | 0.954949 |

| Location | 22,452,014 – 22,452,121 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22452014 107 - 23771897 GACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUGAGGCGCGAGCAUCUAUC---CAAA----AAGCGA-----U (.((((((((((.((((.((((..((......((((...-...)))).))..)))))))).)))))))))).)......((.((....)).))....---....----......-----. ( -31.10) >DroVir_CAF1 40514 109 - 1 GUUCUUAGUUUUAGACGUGUUUUGGUGUGCUUG-AAGUU-UUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCGAUUGUGAGGUAAAAGCGUCCAAAUACUAAA----ACAAAA-----U (..(((((((((.((((.((((..((..((...-.....-..))....))..)))))))).)))))))))..)..(((.((.((....)).)))))........----......-----. ( -26.50) >DroPse_CAF1 16835 108 - 1 GACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUGCGGCAUGAGCAUCUAAC---CAAA----AAAAAA-----U (.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))).)....((((.......)))).....---....----......-----. ( -29.60) >DroMoj_CAF1 38426 118 - 1 GAUCUUAGUUUUAGACGUGUUUUGGUGUGCUUGGGAGUU-UUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCAAUUGUGAGGCGAAAGCAUCCAGAAG-GAAAACAGACAAAAGACCAA (..(((((((((.((((.((((..((.....(((((...-...)))))))..)))))))).)))))))))..)..((((...((....)).(((....)-))......))))........ ( -34.40) >DroAna_CAF1 3060 96 - 1 GCCCUUAGUUUUCGACGUGUUUUGGUGUGCAGGGGAGUU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUGAGGCGUGAGCAUCUA-------AA---------------- (.((((((((((.((((.((((..((......((((...-...)))).))..)))))))).)))))))))).)......((.((....)).))..-------..---------------- ( -31.60) >DroPer_CAF1 16867 108 - 1 GACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUGCGGCAUGAGCAUCUAAC---CAAA----AAAAAA-----U (.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))).)....((((.......)))).....---....----......-----. ( -29.60) >consensus GACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUU_AUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUGAGGCAUGAGCAUCUAAC___CAAA____AAAAAA_____U (.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))).).........((....)).............................. (-26.47 = -26.22 + -0.25)
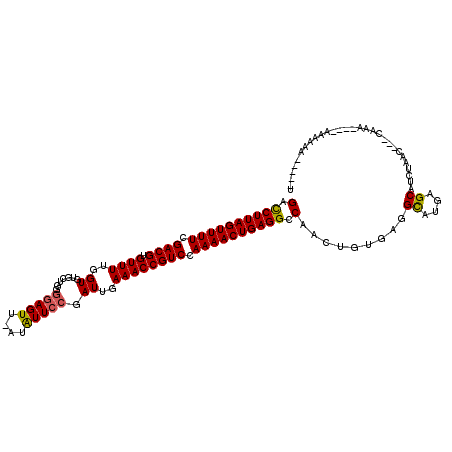
| Location | 22,452,042 – 22,452,145 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22452042 103 + 23771897 CACAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AACUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCUAUUGUGGACAA------------CACU----ACGGAUAU ((((((.(((((.((((((.((((((((.....(((....-...)))...(.....).)))).)))).)))))).))))).)))))).....------------..(.----...).... ( -28.80) >DroVir_CAF1 40545 106 + 1 CACAAUCGGCCUCAGUUUUGGACGGUUUCAAUCGGAACAA-AACUU-CAAGCACACCAAAACACGUCUAAAACUAAGAACCAUUGUGGGCAU------------UAUCUAGUUAGGAAAU ((((((.(..((.((((((((((((((((....)))))..-..(..-...)............))))))))))).))..).)))))).....------------................ ( -24.60) >DroPse_CAF1 16863 96 + 1 CACAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAUCAACUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAG------------UAUU------------ ((((((.(((((.((((((.((((((((.....(((........)))...(.....).)))).)))).)))))).))))).)))))).....------------....------------ ( -28.80) >DroMoj_CAF1 38465 107 + 1 CACAAUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAACAA-AACUCCCAAGCACACCAAAACACGUCUAAAACUAAGAUCCAUUGUGGGCAU------------UUUGUGGGAAGAAAAC ((((((.(..((.((((((((((((((((....)))))..-..((....))............))))))))))).))..).)))))).....------------................ ( -26.00) >DroAna_CAF1 3077 115 + 1 CACAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAG-AACUCCCCUGCACACCAAAACACGUCGAAAACUAAGGGCCAUUGUAGGCAGUAUAUAUUACUAUAUU----ACGAAUCU ......((((((.((((((.((((((((((...(((....-...)))..)).......)))).)))).))))))..))))))((((((..((((.....))))...))----)))).... ( -29.41) >DroPer_CAF1 16895 96 + 1 CACAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAUCAACUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAG------------UAUA------------ ((((((.(((((.((((((.((((((((.....(((........)))...(.....).)))).)))).)))))).))))).)))))).....------------....------------ ( -28.80) >consensus CACAGUUGGCCUCAGUUUUGGACGGUUUCAAUCGGAAUAA_AACUCCCCAGCACACCAAAACACGUCGAAAACUAAGGUCCAUUGUGGACAG____________UAUU____A_G_A_AU ((((((.(((((.((((((.(((((((((....))))).....(......)............)))).)))))).))))).))))))................................. (-23.39 = -23.78 + 0.39)
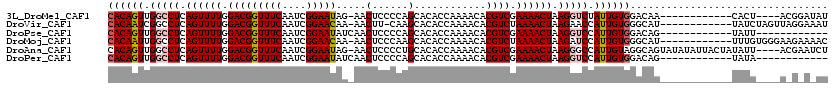

| Location | 22,452,042 – 22,452,145 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22452042 103 - 23771897 AUAUCCGU----AGUG------------UUGUCCACAAUAGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUG .....(((----((((------------(((....)))).(.((((((((((.((((.((((..((......((((...-...)))).))..)))))))).)))))))))).).)))))) ( -29.00) >DroVir_CAF1 40545 106 - 1 AUUUCCUAACUAGAUA------------AUGCCCACAAUGGUUCUUAGUUUUAGACGUGUUUUGGUGUGCUUG-AAGUU-UUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCGAUUGUG ................------------.....((((((.(..(((((((((.((((.((((..((..((...-.....-..))....))..)))))))).)))))))))..).)))))) ( -30.60) >DroPse_CAF1 16863 96 - 1 ------------AAUA------------CUGUCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUG ------------....------------.....((((.(((.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))))))..)))) ( -33.30) >DroMoj_CAF1 38465 107 - 1 GUUUUCUUCCCACAAA------------AUGCCCACAAUGGAUCUUAGUUUUAGACGUGUUUUGGUGUGCUUGGGAGUU-UUGUUCCGAUUGAAACCGUCCAAAACUGAGGCCAAUUGUG ................------------.....((((((((.((((((((((.((((.((((..((.....(((((...-...)))))))..)))))))).)))))))))))).)))))) ( -32.90) >DroAna_CAF1 3077 115 - 1 AGAUUCGU----AAUAUAGUAAUAUAUACUGCCUACAAUGGCCCUUAGUUUUCGACGUGUUUUGGUGUGCAGGGGAGUU-CUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUG ......((----(.((((....)))))))....((((.(((((.((((((((.((((.((((..((......((((...-...)))).))..)))))))).)))))))))))))..)))) ( -34.00) >DroPer_CAF1 16895 96 - 1 ------------UAUA------------CUGUCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUUGAUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUG ------------....------------.....((((.(((.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))))))..)))) ( -33.30) >consensus AU_U_C_U____AAUA____________CUGCCCACAAUGGACCUUAGUUUUCGACGUGUUUUGGUGUGCUGGGGAGUU_AUAUUCCGAUUGAAACCGUCCAAAACUGAGGCCAACUGUG .................................((((.(((.((((((((((.((((.((((..((.......(((((....))))).))..)))))))).)))))))))))))..)))) (-28.70 = -28.48 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:23 2006