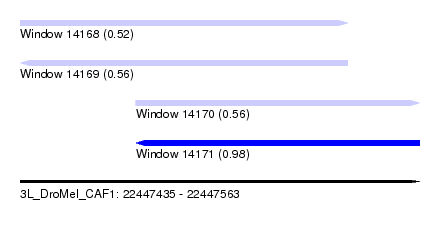
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,447,435 – 22,447,563 |
| Length | 128 |
| Max. P | 0.982000 |

| Location | 22,447,435 – 22,447,540 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.36 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22447435 105 + 23771897 GAACUUUUCAGUUGGCUUUCUUCUGCUCCGGUGUCCCCCGCAACUCCUUUUUUUGAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUU ..........((.(((.....((((((((((......)))..............))).))))((((((.........)))))).........))).))....... ( -24.13) >DroEre_CAF1 3073 105 + 1 GAACUUUUCAGUUGGCUUUCUUCUGCUCCAAUGUCCCCCGCACCACCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUU ((((..(.(((((((((..............(((.....))).............)))))).((((((.........))))))...........))).)..)))) ( -19.13) >DroYak_CAF1 2439 105 + 1 AAACUUUUCAGUUGGCUUUCUUCUACUCCAAUGUCCCCCGCACCUCCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUU ((((..(.(((((((((..............(((.....))).............)))))).((((((.........))))))...........))).)..)))) ( -18.73) >consensus GAACUUUUCAGUUGGCUUUCUUCUGCUCCAAUGUCCCCCGCACCUCCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUU ((((..(.(((((((((..............(((.....))).............)))))).((((((.........))))))...........))).)..)))) (-19.58 = -19.36 + -0.22)

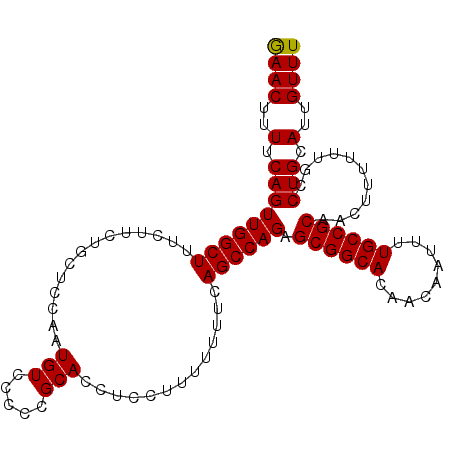
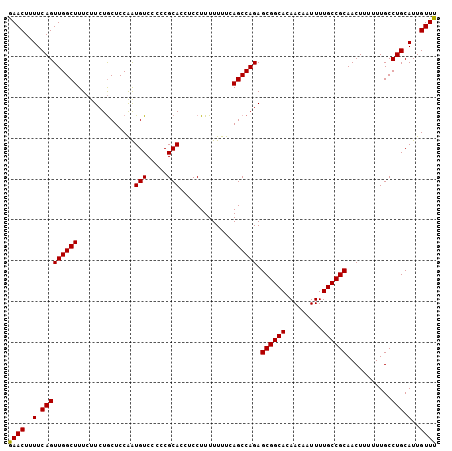
| Location | 22,447,435 – 22,447,540 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -23.33 |
| Energy contribution | -22.89 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22447435 105 - 23771897 AAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUCAAAAAAAGGAGUUGCGGGGGACACCGGAGCAGAAGAAAGCCAACUGAAAAGUUC ..........(((......(..((((((....))))))..)((((((((((........))).....(....).)))))))........)))((((....)))). ( -29.10) >DroEre_CAF1 3073 105 - 1 AAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGUGGUGCGGGGGACAUUGGAGCAGAAGAAAGCCAACUGAAAAGUUC ..........(((.......((((..(((...((((.(.((((.(((.((.......)).))).))))).)))))))..))))......)))((((....)))). ( -26.52) >DroYak_CAF1 2439 105 - 1 AAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGAGGUGCGGGGGACAUUGGAGUAGAAGAAAGCCAACUGAAAAGUUU ..........(((.......((((..(((...((((.(.((((.((..((.......))..)).))))).)))))))..))))......)))((((....)))). ( -23.12) >consensus AAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGAGGUGCGGGGGACAUUGGAGCAGAAGAAAGCCAACUGAAAAGUUC ..........(((......(..((((((....))))))..)((((((((...............)).(....)..))))))........)))((((....)))). (-23.33 = -22.89 + -0.44)

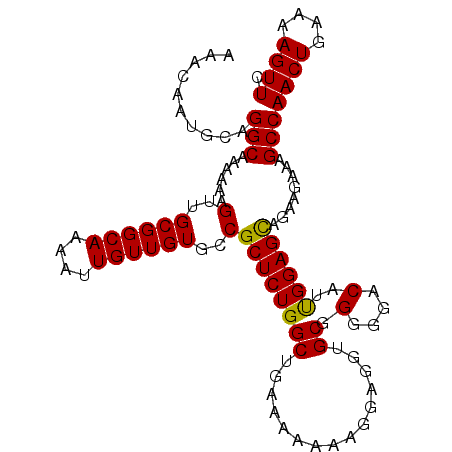
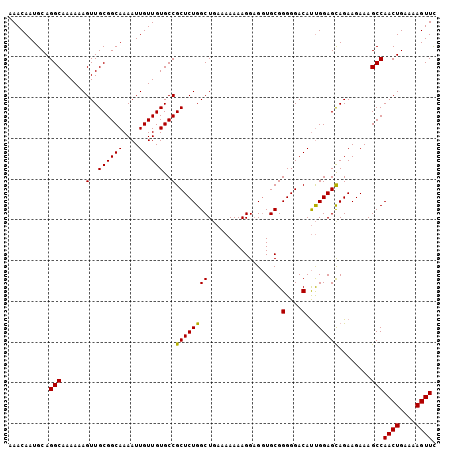
| Location | 22,447,472 – 22,447,563 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22447472 91 + 23771897 CCGCAACUCCUUUUUUUGAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUUUUGACUGCAACGCUCCGGCCGGC ...............(((.(((.(((((((((((.(((..(((.((((.....))))..)))..))).)))..)))..))))).)))))). ( -24.70) >DroEre_CAF1 3110 91 + 1 CCGCACCACCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUUUUGACUGCAACGCUCCGGCCGGC ................((.(((.(((((((((((.(((..(((.((((.....))))..)))..))).)))..)))..))))).))).)). ( -24.40) >DroYak_CAF1 2476 91 + 1 CCGCACCUCCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUUUUGACUGCAACGCUCCGACCGGC ...................(((.(((((((((((.(((..(((.((((.....))))..)))..))).)))..)))..))))).....))) ( -19.70) >consensus CCGCACCUCCUUUUUUUCAGCCAGAGCGGCACAACAAUUUUGCCGCAACUUUUUUGCCUGCAUUGUUUUUGACUGCAACGCUCCGGCCGGC ...................(((.(((((((((((.(((..(((.((((.....))))..)))..))).)))..)))..))))).))).... (-21.53 = -21.87 + 0.33)


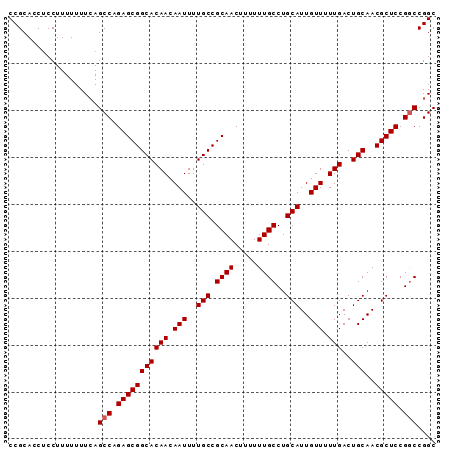
| Location | 22,447,472 – 22,447,563 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -30.29 |
| Energy contribution | -30.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22447472 91 - 23771897 GCCGGCCGGAGCGUUGCAGUCAAAAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUCAAAAAAAGGAGUUGCGG .((((((((((((..((((.(((......(((..((((.....)))).)))...)))))))..)))))))))((........))....))) ( -30.10) >DroEre_CAF1 3110 91 - 1 GCCGGCCGGAGCGUUGCAGUCAAAAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGUGGUGCGG ..(((((((((((..((((.(((......(((..((((.....)))).)))...)))))))..)))))))))))................. ( -32.40) >DroYak_CAF1 2476 91 - 1 GCCGGUCGGAGCGUUGCAGUCAAAAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGAGGUGCGG ..(((((((((((..((((.(((......(((..((((.....)))).)))...)))))))..)))))))))))................. ( -29.70) >consensus GCCGGCCGGAGCGUUGCAGUCAAAAACAAUGCAGGCAAAAAAGUUGCGGCAAAAUUGUUGUGCCGCUCUGGCUGAAAAAAAGGAGGUGCGG ..(((((((((((..((((.(((......(((..((((.....)))).)))...)))))))..)))))))))))................. (-30.29 = -30.40 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:17 2006