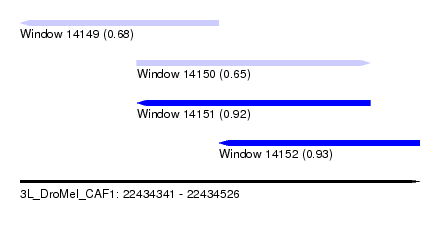
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,434,341 – 22,434,526 |
| Length | 185 |
| Max. P | 0.929823 |

| Location | 22,434,341 – 22,434,433 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434341 92 - 23771897 --UGUUGCUCUUACUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGAGCGAAUGGCAGAUACAACAGCCGC-------------------------- --(((((......(((((((...(((((.......((.(((.......)))))........(....)..))))))))))))...))))).....-------------------------- ( -21.50) >DroPse_CAF1 4158 120 - 1 GCUGUUGCUGUUGCUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGACCGAAUGGCAGAUACAGCUACAGCAACUGCCCCUCCACCAUCCCCCCGGC (((((.(((((..(((((((.....(((.......((.(((.......))))).......)))...(.....).)))))))..))))).)))))....(((................))) ( -33.83) >DroSim_CAF1 11185 92 - 1 --UGUUGCUGUUACUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGAGCGAAUGGCAGAUACAACAGCCGC-------------------------- --.((.((((((.(((((((...(((((.......((.(((.......)))))........(....)..))))))))))))....)))))).))-------------------------- ( -25.90) >DroPer_CAF1 4231 120 - 1 GCUGUUGCUGUUGCUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGACCGAAUGGCAGAUACAGCUACAGCAACUGCCCCUCCACCAUCCCCCCGGC (((((.(((((..(((((((.....(((.......((.(((.......))))).......)))...(.....).)))))))..))))).)))))....(((................))) ( -33.83) >DroEre_CAF1 12114 92 - 1 --UGUUGCUCUCACUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGAGCGAAUGGAAGAUACAACAGCCGC-------------------------- --..(((((((...(((........((((.....))))(((.......)))..........)))...)))))))..((..(......)..))..-------------------------- ( -16.10) >DroYak_CAF1 48544 92 - 1 --UGUUGUUUUUACUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGAGCGAAUGGCAGAUACAACAGCCGC-------------------------- --((((((.....(((((((...(((((.......((.(((.......)))))........(....)..))))))))))))..)))))).....-------------------------- ( -23.90) >consensus __UGUUGCUGUUACUGCCAUAUUUCGCUGCACACCGGCUGCCAUUUCCGCACCAAACAAAAGCAAACAGAGCGAAUGGCAGAUACAACAGCCGC__________________________ ...((((......(((((((...(((((.......((.(((.......)))))........(....)..))))))))))))...))))................................ (-17.97 = -18.25 + 0.28)


| Location | 22,434,395 – 22,434,503 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434395 108 + 23771897 CAGCCGGUGUGCAGCGAAAUAUGGCAGUAAGAGCAACA---------ACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAA--- ..((..((((((.(((..(((((((.(((...((....---------.......)).)))))))))))))((((...........))))))))))((((........))))..))..--- ( -28.30) >DroPse_CAF1 4238 120 + 1 CAGCCGGUGUGCAGCGAAAUAUGGCAGCAACAGCAACAGCAAAAACGACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAAAAG ..((((....(((((.....(((((.((((((((....))..............((....))..))))))))))).....)))))))))......((((........))))......... ( -30.40) >DroSim_CAF1 11239 108 + 1 CAGCCGGUGUGCAGCGAAAUAUGGCAGUAACAGCAACA---------ACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAA--- ..((..((((((.(((..(((((((.(((...((....---------.......)).)))))))))))))((((...........))))))))))((((........))))..))..--- ( -28.30) >DroPer_CAF1 4311 120 + 1 CAGCCGGUGUGCAGCGAAAUAUGGCAGCAACAGCAACAGCAAAAACGACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAAAAG ..((((....(((((.....(((((.((((((((....))..............((....))..))))))))))).....)))))))))......((((........))))......... ( -30.40) >DroEre_CAF1 12168 108 + 1 CAGCCGGUGUGCAGCGAAAUAUGGCAGUGAGAGCAACA---------ACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAA--- ..((..((((((.(((..(((((((.((((........---------...))))(....)))))))))))((((...........))))))))))((((........))))..))..--- ( -29.30) >DroYak_CAF1 48598 108 + 1 CAGCCGGUGUGCAGCGAAAUAUGGCAGUAAAAACAACA---------ACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAA--- ..((..((((((.(((..(((((((.(((.........---------..........)))))))))))))((((...........))))))))))((((........))))..))..--- ( -26.51) >consensus CAGCCGGUGUGCAGCGAAAUAUGGCAGUAACAGCAACA_________ACAUCACGCAUACGCCAUGUUGCGCCAUUUGAAGUUGUUGGCGUAUACGUAAUUUUUAUUUUACAAGCAA___ ..((..((((((.(((..(((((((.((....))....................(....)))))))))))((((...........))))))))))((((........))))..))..... (-26.49 = -26.55 + 0.06)

| Location | 22,434,395 – 22,434,503 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -27.19 |
| Energy contribution | -27.05 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434395 108 - 23771897 ---UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU---------UGUUGCUCUUACUGCCAUAUUUCGCUGCACACCGGCUG ---..((..(((((((((..(((((((((((((((...........((......))))))))))))))))).)---------))))....)))).))........((((.....)))).. ( -27.70) >DroPse_CAF1 4238 120 - 1 CUUUUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGUCGUUUUUGCUGUUGCUGUUGCUGCCAUAUUUCGCUGCACACCGGCUG .........((((((.....(((((((((((((((...........((......))))))))))))))))).....)))))).((((.(((.((.((........)).))))).)))).. ( -34.50) >DroSim_CAF1 11239 108 - 1 ---UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU---------UGUUGCUGUUACUGCCAUAUUUCGCUGCACACCGGCUG ---..((..((((..(((..(((((((((((((((...........((......)))))))))))))))))..---------..)))...)))).))........((((.....)))).. ( -28.40) >DroPer_CAF1 4311 120 - 1 CUUUUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGUCGUUUUUGCUGUUGCUGUUGCUGCCAUAUUUCGCUGCACACCGGCUG .........((((((.....(((((((((((((((...........((......))))))))))))))))).....)))))).((((.(((.((.((........)).))))).)))).. ( -34.50) >DroEre_CAF1 12168 108 - 1 ---UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU---------UGUUGCUCUCACUGCCAUAUUUCGCUGCACACCGGCUG ---.(((..(..(((((...(((((((((((((((...........((......)))))))))))))))))..---------....((.......))..)))))..).)))......... ( -27.40) >DroYak_CAF1 48598 108 - 1 ---UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU---------UGUUGUUUUUACUGCCAUAUUUCGCUGCACACCGGCUG ---..((..(((((((((..(((((((((((((((...........((......)))))))))))))))))..---------..))).)))))).))........((((.....)))).. ( -30.40) >consensus ___UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU_________UGUUGCUGUUACUGCCAUAUUUCGCUGCACACCGGCUG ....(((..(..(((((...(((((((((((((((...........((......)))))))))))))))))...............((.......))..)))))..).)))......... (-27.19 = -27.05 + -0.14)
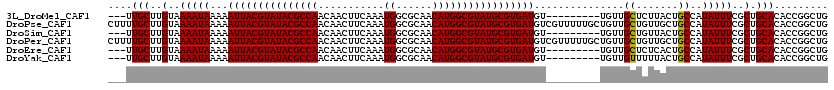


| Location | 22,434,433 – 22,434,526 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434433 93 - 23771897 UAUUCCCGUU--------------UUUUCGUUUUUGU------UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU------- ..........--------------.......((((((------((.......))))))))(((((((((((((((...........((......)))))))))))))))))..------- ( -23.80) >DroSec_CAF1 6287 93 - 1 UAUUCCCGUU--------------UUUUCGUUUUUGU------UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU------- ..........--------------.......((((((------((.......))))))))(((((((((((((((...........((......)))))))))))))))))..------- ( -23.80) >DroSim_CAF1 11277 93 - 1 UAUUCCCGUU--------------UUUUCGUUUUUGU------UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU------- ..........--------------.......((((((------((.......))))))))(((((((((((((((...........((......)))))))))))))))))..------- ( -23.80) >DroPer_CAF1 4351 106 - 1 UCUUUCCGUU--------------UUUUAGUUUUUGUUCGCUUUUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGUCGUUUUU .......(((--------------((..(((....(....)....)))...)))))....(((((((((((((((...........((......)))))))))))))))))......... ( -22.80) >DroEre_CAF1 12206 107 - 1 UAUUCGCGUUUUCGCGUUUUCGUUUUUUCGUUUUUGU------UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU------- ....((((....))))...............((((((------((.......))))))))(((((((((((((((...........((......)))))))))))))))))..------- ( -28.60) >DroYak_CAF1 48636 92 - 1 UAUUCCCGUU---------------UUUCGUUUUUGU------UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU------- ..........---------------......((((((------((.......))))))))(((((((((((((((...........((......)))))))))))))))))..------- ( -23.80) >consensus UAUUCCCGUU______________UUUUCGUUUUUGU______UUGCUUGUAAAAUAAAAAUUACGUAUACGCCAACAACUUCAAAUGGCGCAACAUGGCGUAUGCGUGAUGU_______ ............................................................(((((((((((((((...........((......)))))))))))))))))......... (-20.90 = -20.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:00 2006