
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,434,147 – 22,434,250 |
| Length | 103 |
| Max. P | 0.834365 |

| Location | 22,434,147 – 22,434,250 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -24.93 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434147 103 + 23771897 GAAUGGAGGGGUGGUGG------------GUGCUGGCGCCUGGGGUGGUGCAGCUGUGGCGGGUUAUGUAUAAAUUCAUAAAGAGACCCACAGCAGCGAGAGCGAGAAGGGAAAA ..........((.((((------------((((((.((((......)))))))).....(...(((((........)))))...))))))).)).((....))............ ( -30.40) >DroEre_CAF1 11910 113 + 1 GG-GGGUCGUGUGGCGGGUGGUGGUGCUGGUGCUGGUGCCCAGGGUGGCGCAGCUGUGGCGGGUUAUGUAUAAAUUCAUAAAGAGACCCACGGCAGCGAGAACGAGAAGGGA-AA ..-..(((((((.((((.((((..(.((((.((....)))))).)..)).)).)))).))(((((...(((......)))....))))))))))..................-.. ( -35.40) >DroYak_CAF1 48344 109 + 1 GGAUGGUGGUGCGGUGGG------UGCUGGUGCUGGUGCCUGGGGUGGUGCAGCUGUGCCGGGUUAUGUAUAAAUUCAUAAAGAGACCCACAGCAGCGAGAACGAGAAGAGAAAA ......((.(((.(((((------(.((((..(..(..((......))..)....)..)))).(((((........)))))....)))))).))).))................. ( -35.70) >consensus GGAUGGUGGUGUGGUGGG______UGCUGGUGCUGGUGCCUGGGGUGGUGCAGCUGUGGCGGGUUAUGUAUAAAUUCAUAAAGAGACCCACAGCAGCGAGAACGAGAAGGGAAAA ..............................((((.(((((......))))).(((((((....(((((........)))))......)))))))))))................. (-24.93 = -24.60 + -0.33)

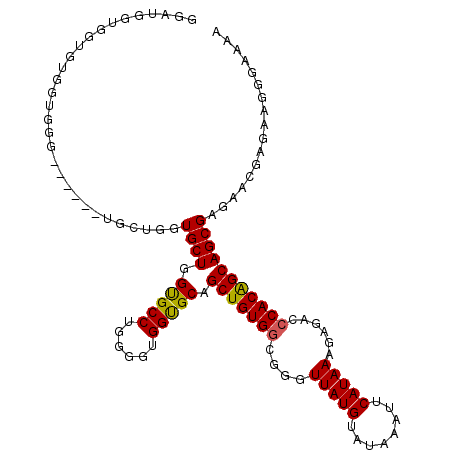

| Location | 22,434,147 – 22,434,250 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22434147 103 - 23771897 UUUUCCCUUCUCGCUCUCGCUGCUGUGGGUCUCUUUAUGAAUUUAUACAUAACCCGCCACAGCUGCACCACCCCAGGCGCCAGCAC------------CCACCACCCCUCCAUUC ............(((...((((..((((((.....((((........))))))))))..)))).((.((......)).)).)))..------------................. ( -22.40) >DroEre_CAF1 11910 113 - 1 UU-UCCCUUCUCGUUCUCGCUGCCGUGGGUCUCUUUAUGAAUUUAUACAUAACCCGCCACAGCUGCGCCACCCUGGGCACCAGCACCAGCACCACCACCCGCCACACGACCC-CC ..-.......((((....((((..((((((.....((((........))))))))))..)))).(((((......)))....((....))..........))...))))...-.. ( -25.40) >DroYak_CAF1 48344 109 - 1 UUUUCUCUUCUCGUUCUCGCUGCUGUGGGUCUCUUUAUGAAUUUAUACAUAACCCGGCACAGCUGCACCACCCCAGGCACCAGCACCAGCA------CCCACCGCACCACCAUCC ..................(.(((.((((((((..(((((........)))))...((....((((..((......))...)))).)))).)------))))).))).)....... ( -22.90) >consensus UUUUCCCUUCUCGUUCUCGCUGCUGUGGGUCUCUUUAUGAAUUUAUACAUAACCCGCCACAGCUGCACCACCCCAGGCACCAGCACCAGCA______CCCACCACACCACCAUCC ............(((...((((..((((((.....((((........))))))))))..))))(((.(.......))))..)))............................... (-15.91 = -15.80 + -0.11)

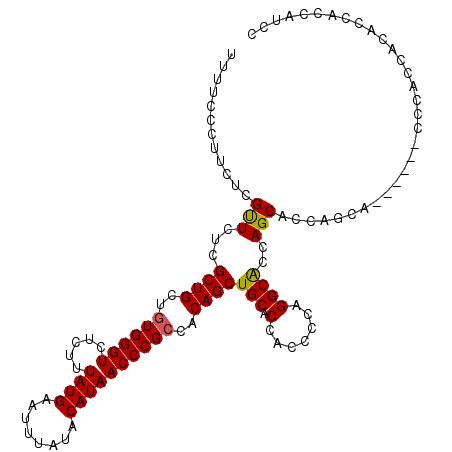

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:56 2006