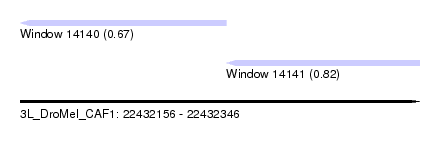
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,432,156 – 22,432,346 |
| Length | 190 |
| Max. P | 0.824187 |

| Location | 22,432,156 – 22,432,254 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22432156 98 - 23771897 UUCACACUGAAAUAGCGCGAAAUA-------GCAAGAGCAGCUAAUGGCGUGUGAAAAGUAAUGGAAAUCAACCCGGAUUUCUAUAAGUUAUGGUAAGAGUGG-AA ((((((((......(((((...((-------((.......))))....)))))....))).(((((((((......)))))))))..............))))-). ( -24.10) >DroPse_CAF1 1878 92 - 1 UUGCCACCGCCA----CCGAAAU--------GUACGAGCAGCUAAUGGCGUGUGGAAAUCAAUGAGAGUCGAAGCGGAAUUUUAUAAUUUAUGGCGG-CGCAA-AA ...(((((((((----......(--------((....))).....))))).)))).......((...((((.....(((((....)))))....)))-).)).-.. ( -22.30) >DroSec_CAF1 4032 98 - 1 UUCACACUGAAAUAGCGCGAAAUA-------GCACGAGCAGCUAAUGGCGUGUGGAAAUUAAUGGAAAUCAACCCGGAUUUCUAUAAGUUACGGUAAAAGUGG-AA (((((((((.....(((((...((-------((.......))))....)))))...((((.(((((((((......))))))))).)))).))))....))))-). ( -25.00) >DroSim_CAF1 9007 98 - 1 UUCACACUGAAAUAGCGCGAAAUA-------GCACGAGCAGCUAAUGGCGUGUGGAAAUUAAUGGAAAUCAACCCGGAUUUCUAUAAGUUACGGUAAGAGUGG-AA (((((((((.....(((((...((-------((.......))))....)))))...((((.(((((((((......))))))))).)))).))))....))))-). ( -25.00) >DroEre_CAF1 10083 106 - 1 UUCGCACUGAAAUAGCGCGAAAUAGCAAAUAGCAGGAGCAGCUAAUGGCGUGUGGAAAUUAAUGGAAAUCAACACGGAUUUCUGCAAGUUACGGUAAGUUCUAUAA (((((.(((...))).)))))...((.....)).(((((.(((..((((.((..((((((..((........))..))))))..)).)))).)))..))))).... ( -27.00) >DroYak_CAF1 45765 99 - 1 UUCACACUGAAAUUGCGCGAAAUA-------GCACGAGCAGCUAAUGGCGUGUGGAAAUUAAUGGAAAUCAACCCGUAUUUCUAUAAGUUACGGUAAGAGUGAAAA (((((.(((...(((.((......-------)).))).)))....((((.(((((((((..((((........))))))))))))).))))........))))).. ( -24.60) >consensus UUCACACUGAAAUAGCGCGAAAUA_______GCACGAGCAGCUAAUGGCGUGUGGAAAUUAAUGGAAAUCAACCCGGAUUUCUAUAAGUUACGGUAAGAGUGA_AA (((((((........................((....)).((.....)))))))))((((.(((((((((......))))))))).))))................ (-16.64 = -17.03 + 0.39)

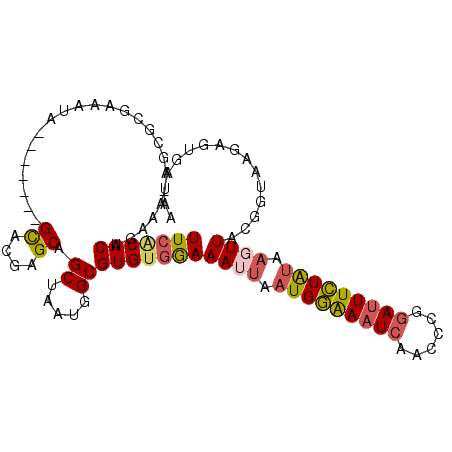
| Location | 22,432,254 – 22,432,346 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.32 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -17.71 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22432254 92 - 23771897 GCACCCUGC--CGACAGAAAGAGGUG-------GGGAUAGCGGUUGCUGUGGUUGCUGGCAGCUUAAGAGUUGUCGGUUAGUUGCCCGAGACCGC---------UUGAGG ...(((..(--(..........))..-------)))..(((((((..((.(((.((((((((((....)))))))))).....))))).))))))---------)..... ( -38.90) >DroPse_CAF1 1970 102 - 1 GCUUGUGGCACGGUGUGGCUGGGGCG-------GCCACCGAGGCAGCUGCGGUGGCUGGCAGUUUAA-AGUUGUCGGUUAGUUGCCUCAGAUCUCGCCGAGGCGUCGCCG ((.....)).(((((.((((((((((-------(((((((((....)).)))))((((((((.....-..))))))))..)))))))))).)).))))).(((...))). ( -46.50) >DroSec_CAF1 4130 92 - 1 GCACCCUGC--UGAGAGGCGAAGGUU-------GGGAUAGCGGUAGCUGUGGUUGCUGGCAGCUUAAGAGUUGUCGUUUAGUUGCCCGCGACCGC---------UUGAGG ((.((((((--(....))))......-------)))(((((....)))))((((((.(((((((...((....))....))))))).))))))))---------...... ( -36.90) >DroSim_CAF1 9105 92 - 1 GCACCCUGC--UGAGGGGCGAAGGUU-------GGGAUAGCGGUAGCUGUGGUUGCUGGCAGCUUAAGAGUUGUCGGUUAGUUGCCCGCGACCGC---------UUGAGG ((.((((..--..)))))).....(.-------.(((((((....)))))((((((.(((((((...((....))....))))))).)))))).)---------)..).. ( -38.00) >DroEre_CAF1 10189 99 - 1 GCACCCUGC--UGAGGGCAGGAGGUGAGGGGCGGGGAUAGCGGUAGCUGUGGUUGCUGGCAGCUUAAGAGUUGUCGGUUAGCUGACCGCGACCGC---------UUGAGG ((..(((((--.....)))))..))...((((((.....((((((((((.....((((((((((....))))))))))))))).)))))..))))---------)).... ( -46.20) >DroYak_CAF1 45864 83 - 1 GCACCAUGC--UGAGGGUAGGCGGUG--------GGCUAGCGGUAGCUAUGGCUGCUGGA--------AGUUGUCGGUUAGUUGCCCGCGAGCCC---------UUGAGG ((.....))--.((((((..((((..--------(((((((((((((....)))))).((--------.....)).)))))))..))))..))))---------)).... ( -33.40) >consensus GCACCCUGC__UGAGGGGAGGAGGUG_______GGGAUAGCGGUAGCUGUGGUUGCUGGCAGCUUAAGAGUUGUCGGUUAGUUGCCCGCGACCGC_________UUGAGG .................................((....((((((((((.....((((((((((....)))))))))))))))).))))..))................. (-17.71 = -19.22 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:50 2006