| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,605,968 – 2,606,058 |
| Length | 90 |
| Max. P | 0.586041 |

| Location | 2,605,968 – 2,606,058 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.55 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
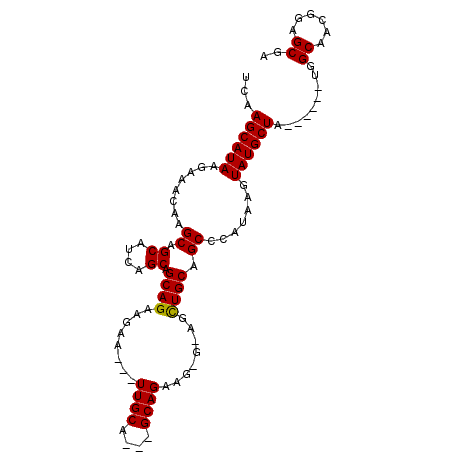
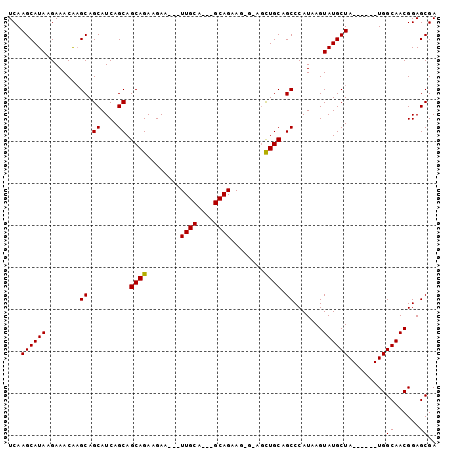
>3L_DroMel_CAF1 2605968 90 - 23771897 UCAAGCAUAAGAAACAAGCAGCAUCAGCAGCAGAAGAA---UUGCA---GCAGAAGCGUAGCUGCAGCCCAUAAGUAUGCUA------UGGCAACGGAGCGA ....((...........(((((....((((........---)))).---((....))...))))).(((.(((.(....)))------))))......)).. ( -20.70) >DroEre_CAF1 4367 73 - 1 UCAAGCAUAAGAAACAAGCAGCAUCAGCAGCAGAAGAA-----------------------UUGCAGCCCAUAAGUAUGCUA------UGGCAACGGAGCGA ...((((((........((.((....)).((((.....-----------------------)))).)).......)))))).------((....))...... ( -12.86) >DroYak_CAF1 7407 101 - 1 UCAAGCAUAAGAAACG-GCAGCAUCAGCAGCAGAAGAAGAAUUGCAGCAGCAGAAGAGGAGCUGCAGCCCAUAAGUAUGCUAUGGCUAUGGCAACGGAGCGA .((((((((......(-((.((....)).((((........)))).(((((.........))))).)))......)))))).))(((.((....)).))).. ( -29.50) >consensus UCAAGCAUAAGAAACAAGCAGCAUCAGCAGCAGAAGAA___UUGCA___GCAGAAG_G_AGCUGCAGCCCAUAAGUAUGCUA______UGGCAACGGAGCGA ...((((((........((.((....)).((((........((((....))))........)))).)).......)))))).........((......)).. (-15.90 = -16.02 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:02 2006