
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,431,323 – 22,431,477 |
| Length | 154 |
| Max. P | 0.725889 |

| Location | 22,431,323 – 22,431,437 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.30 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -36.46 |
| Energy contribution | -36.10 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22431323 114 - 23771897 AUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCGCCAUUUGCAGGAUUUGCGCCUGCAAUGGGCGG-GCA ......(((((.......)))))..(((((((......(((......(((((....(....)..))))).......)))(((((.(((((.......))))))))))))))-))) ( -39.82) >DroSec_CAF1 3040 115 - 1 AUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCGCCAUUUGCAGGAUUUGCGCCUGAGAUGGGCGGGGUA ............(((((((((((((..(((((......(((......(((((....(....)..))))).......)))......)))))..)))))))........)))))).. ( -37.02) >DroSim_CAF1 8171 115 - 1 AUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCGCCAUUUGCAGGAUUUGCGCCUGAGAUGGGCGGGGUA ............(((((((((((((..(((((......(((......(((((....(....)..))))).......)))......)))))..)))))))........)))))).. ( -37.02) >DroEre_CAF1 9256 114 - 1 AUGUAAGCACUUCUCCGCGGUGCGGUGCCUGCUUUCUCCGCUUUCGCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCGCCAUUUGCAGGAUUUGCGCCUGCGAUGGGCGGAGC- ............(((((((((((.(((..((((((((..((....))(((.......)))))).))))).)))...)))))..(((((((.......)))))))...)))))).- ( -40.80) >DroYak_CAF1 44928 115 - 1 AUGUAAGCACUUCUCCGCGGUGCGGUCCCUGCUUUCUACGUUUUCUCUGCUUUCUCCGCAAGAUAAGCACCACAAUGCGCCAUUUGCAGGAUUUGCGCCUGCGAUGGGCGGAGUA ............(((((((((((((..(((((......(((......(((((....(....)..))))).......)))......)))))..)))))))..(....))))))).. ( -37.12) >consensus AUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCGCCAUUUGCAGGAUUUGCGCCUGCGAUGGGCGGAGUA ............(((((((((((((..(((((......(((......(((((....(....)..))))).......)))......)))))..)))))))........)))))).. (-36.46 = -36.10 + -0.36)

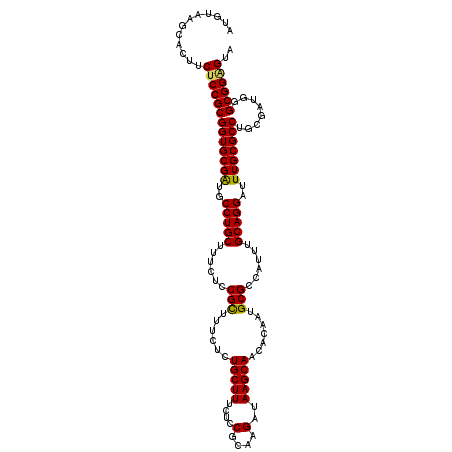

| Location | 22,431,358 – 22,431,477 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.68 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22431358 119 + 23771897 CGCAUUGUGUUGCUUAUCUUGCGGAGAAAGCAGAGAAAGCGGAGAAAGCAGGCAUCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGCAAACAAGGAC .((.(((((((((((.((((((.......))).))))))).......((..(((((((...))))....((((........))))...))).))......))))))))).......... ( -31.40) >DroSec_CAF1 3076 119 + 1 CGCAUUGUGUUGCUUAUCUUGCGGAGAAAGCAGAGAAAGCGGAGAAAGCAGGCAUCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGUAAACAAGGAC ....(((((((((...((((((.......))).)))..(((.((((((((..(..(((...)))..)...)))))...))).)))....)))))))))..................... ( -30.40) >DroSim_CAF1 8207 119 + 1 CGCAUUGUGUUGCUUAUCUUGCGGAGAAAGCAGAGAAAGCGGAGAAAGCAGGCAUCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGCAAACAAGGAC .((.(((((((((((.((((((.......))).))))))).......((..(((((((...))))....((((........))))...))).))......))))))))).......... ( -31.40) >DroEre_CAF1 9291 119 + 1 CGCAUUGUGUUGCUUAUCUUGCGGAGAAAGCAGCGAAAGCGGAGAAAGCAGGCACCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGCAAACAAGGAC .((.(((((((((((.((((((.......)).((....)).)))))))).....((((...))))....(((((..(((((....))))).)))))....))))))))).......... ( -36.90) >DroYak_CAF1 44964 119 + 1 CGCAUUGUGGUGCUUAUCUUGCGGAGAAAGCAGAGAAAACGUAGAAAGCAGGGACCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGCAAACAAGGAC .((.(((((((((...((((((.........................))))))...)))))))))....(((((..(((((....))))).)))))...........)).......... ( -30.81) >consensus CGCAUUGUGUUGCUUAUCUUGCGGAGAAAGCAGAGAAAGCGGAGAAAGCAGGCAUCGCACCGCGGAGAAGUGCUUACAUUUGCGCAAAUGCAGCACAACAAACACAAGCAAACAAGGAC .((.(((((((((((.((((((.......)))).)))))).......((..(((((((...))))....((((........))))...))).))......))))))))).......... (-28.68 = -28.88 + 0.20)

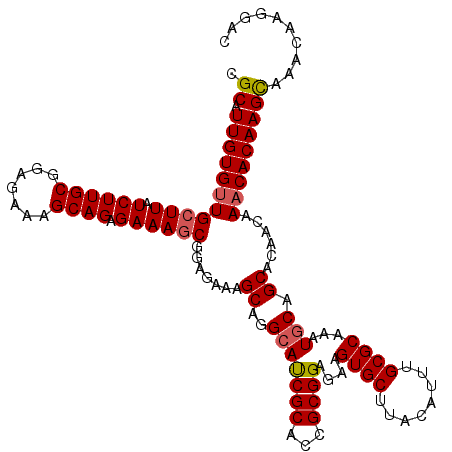
| Location | 22,431,358 – 22,431,477 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22431358 119 - 23771897 GUCCUUGUUUGCUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCG ..........(((((((((....(((((((((((....)))))).)))))......((((.((...)))))).......(((((((.(((.......)))))).))))))))))).)). ( -33.70) >DroSec_CAF1 3076 119 - 1 GUCCUUGUUUACUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCG ....(((((((((((((......(((((((((((....)))))).)))))......((((.(((.....))).....))))...............)))))).)))))))......... ( -31.10) >DroSim_CAF1 8207 119 - 1 GUCCUUGUUUGCUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCG ..........(((((((((....(((((((((((....)))))).)))))......((((.((...)))))).......(((((((.(((.......)))))).))))))))))).)). ( -33.70) >DroEre_CAF1 9291 119 - 1 GUCCUUGUUUGCUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGGUGCCUGCUUUCUCCGCUUUCGCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCG ..........(((((((((.((((((((((((((....)))))).))))).(((..((((.((((((...((.......))...)))))).....)))).)))..)))))))))).)). ( -39.60) >DroYak_CAF1 44964 119 - 1 GUCCUUGUUUGCUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGGUCCCUGCUUUCUACGUUUUCUCUGCUUUCUCCGCAAGAUAAGCACCACAAUGCG ......(((((...((((((((((((((((((((....)))))).)))))......((((.((((.....((.......)).....)))).....))))..)))))))))..))).)). ( -32.50) >consensus GUCCUUGUUUGCUUGUGUUUGUUGUGCUGCAUUUGCGCAAAUGUAAGCACUUCUCCGCGGUGCGAUGCCUGCUUUCUCCGCUUUCUCUGCUUUCUCCGCAAGAUAAGCAACACAAUGCG ((..((((((((((((.((....(((((((((((....)))))).)))))......((((.(((......((.......))......))).....)))))).)))))))).)))).)). (-29.98 = -30.22 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:46 2006