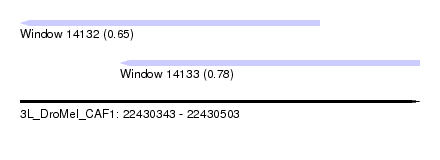
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,430,343 – 22,430,503 |
| Length | 160 |
| Max. P | 0.775096 |

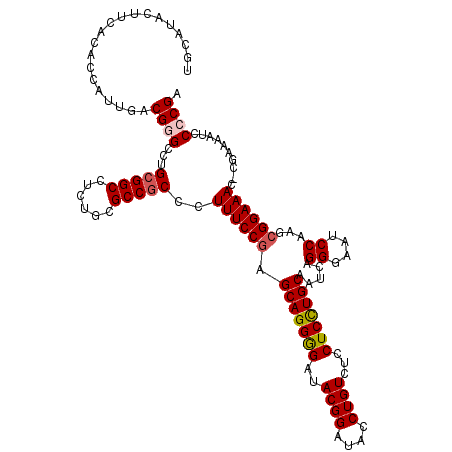
| Location | 22,430,343 – 22,430,463 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.14 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.59 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22430343 120 - 23771897 CGCCCUUUCCAAGCAGGGGAUACGGAUACCUGUAUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAACGCGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGGUGCGGCGCGUCGGA (((((...(((.(((((((((((((....)))))))).)))))..((.((((......(((....))).....)))).)).......................)))...)).)))..... ( -45.40) >DroSec_CAF1 2045 119 - 1 CGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGGUGCGGCGCGGCGCA ((((.((((((.((((((((.((((....)))).))).))))).....((....))...)))))).-.........(((.((((.................))))...)))...)))).. ( -37.43) >DroSim_CAF1 7177 119 - 1 CGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCUUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGGUGCGGCGCGGCGCA ((((....(((.((.((((((..((((.((((....(....)....))))..))))...(((...)-))...))))))..((((.................)))))).)))...)))).. ( -34.73) >DroEre_CAF1 8379 119 - 1 CGCCCUUUCCGAGCAGGAGAUACGGCUACCUGUCUGCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGGUGCGGCGCGGCGAA ((((.((((((.(((((((..((((....))))...))))))).....((....))...)))))).-.........(((.((((.................))))...)))...)))).. ( -36.43) >DroYak_CAF1 44006 114 - 1 CGCCCUUUCCGAGCAGGAGUUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGCUGCG-----GCGAA ((((.((((((((((((((..((((....))))...)))))))..)).))))).....((((....-((........)).((((.................)))))))))-----))).. ( -31.83) >consensus CGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC_CGAAAAUCCCCGACACUUUAUUUUAUUACAUUUUAGUGGUGCGGCGCGGCGAA ((((.((((((.(((((((..((((....))))...))))))).....((....))...))))))...........(((.((((.................))))...)))...)))).. (-30.19 = -30.59 + 0.40)

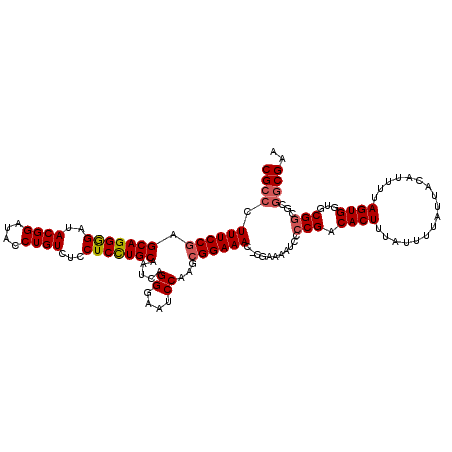
| Location | 22,430,383 – 22,430,503 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -32.94 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22430383 120 - 23771897 UGCAUACUUCACACCAUUGACGGGCCUGCGGCUUCUGCGCCGCCCUUUCCAAGCAGGGGAUACGGAUACCUGUAUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAACGCGAAAAUCCCCGA ....................((((((((((((......))))).........(((((((((((((....)))))))).)))))....)))........(((....))).......)))). ( -48.30) >DroSec_CAF1 2085 119 - 1 UGCAUACUUCACACCAUUGACGGGCCUGCGGCCUCUGCGCCGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGA ....................((((...(((((......)))))..((((((.((((((((.((((....)))).))).))))).....((....))...)))))).-........)))). ( -40.90) >DroSim_CAF1 7217 119 - 1 UGCAUACUUCACACCAUUGACGGGCCUGCGGCCUCUGCGCCGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCUUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGA ....................((((...(((((......)))))..((((((.((((((((.((((....)))).))).))))).....((....))...)))))).-........)))). ( -38.20) >DroEre_CAF1 8419 119 - 1 UGCAUACUUCACACCAUUGACGCGCCCGAGGCCUCGGCGCCGCCCUUUCCGAGCAGGAGAUACGGCUACCUGUCUGCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGA .((.....(((......))).(((((.((....))))))).))..((((((.(((((((..((((....))))...))))))).....((....))...)))))).-............. ( -35.00) >DroYak_CAF1 44041 119 - 1 UGCAUACUUCACACCAUUGGCGUGCUUGCGGCCUCUGCGCCGCCCUUUCCGAGCAGGAGUUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC-CGAAAAUCCCCGA .(((..((..........))..)))..(((((......)))))..((((((.(((((((..((((....))))...))))))).....((....))...)))))).-............. ( -35.80) >consensus UGCAUACUUCACACCAUUGACGGGCCUGCGGCCUCUGCGCCGCCCUUUCCGAGCAGGGGAUACGGAUACCUGUCUCCUCCUGCAAUCAGGGAAUCCAAGCGGAAAC_CGAAAAUCCCCGA ....................((((...(((((......)))))..((((((.(((((((..((((....))))...))))))).....((....))...))))))..........)))). (-32.94 = -33.34 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:41 2006