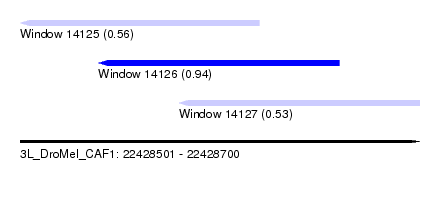
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,428,501 – 22,428,700 |
| Length | 199 |
| Max. P | 0.943620 |

| Location | 22,428,501 – 22,428,620 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -37.75 |
| Energy contribution | -37.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22428501 119 - 23771897 UAAAUUUGCAUUUAAAUGUGCCAUGGCGAACGGCCCGGGGAACGGGCGGCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCGGCCUCAGGAGGGCCUGAAAAAGAGCUGGAAUUAUAAUAA ..(((((((((......)))).........(.(((((.....))))).)((((((((((.((((..((((.((((....))))))))..)))).)))...))))))))))))....... ( -41.40) >DroSec_CAF1 203 119 - 1 UAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCGGCCUCAGGAGGGCCUGAAAAAGAGCUGGAAUUAUAAUAA ..(((((((((......))))...(((((((((((((((((((.(....).))))))))..((.....)).))))))))(((((.....))))).........))).)))))....... ( -40.20) >DroSim_CAF1 5109 119 - 1 UAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCGGCCUCAGGAGGGCCUGAAAAAGAGCUGGAAUUAUAAUAA ..(((((((((......))))...(((((((((((((((((((.(....).))))))))..((.....)).))))))))(((((.....))))).........))).)))))....... ( -40.20) >DroYak_CAF1 40298 119 - 1 UAAAUUUGCAUUCAAAUGUGUCAUGGCGAAUGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCGGCCUGAGAAGGGCUUGAAAAAGAGCUGGAAUUAUAAUAG ...((((((...((....)).....)))))).(((((.....))))).(((((((((((.((((..((..(((((....)))))..)).)))).)))...))))))))........... ( -39.00) >consensus UAAAUUUGCAUUCAAAUGUGUCAUGGCGAACGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCGGCCUCAGGAGGGCCUGAAAAAGAGCUGGAAUUAUAAUAA ....(((((...((....)).....)))))..(((((.....))))).(((((((((((.((((..((((.((((....))))))))..)))).)))...))))))))........... (-37.75 = -37.62 + -0.12)


| Location | 22,428,540 – 22,428,660 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.50 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -32.84 |
| Energy contribution | -34.52 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22428540 120 - 23771897 CCCGUUGGUUUUUGUUGUUAGCACAUUGACACGUUGUGCAUAAAUUUGCAUUUAAAUGUGCCAUGGCGAACGGCCCGGGGAACGGGCGGCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCG ((((..(((((((((..(..(((((((........(((((......)))))...))))))).)..))))).))))))))...((((((((....(((((........))))))))))))) ( -45.40) >DroSec_CAF1 242 120 - 1 CCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCG ....................((.(((.(((((((((((((......))))...))))))))))))))((((((((((((((((.(....).))))))))..((.....)).)))))))). ( -45.80) >DroSim_CAF1 5148 120 - 1 CCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCG ....................((.(((.(((((((((((((......))))...))))))))))))))((((((((((((((((.(....).))))))))..((.....)).)))))))). ( -45.80) >DroEre_CAF1 6310 99 - 1 CCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAACGCUGCAGGGAACGGGCGCCGCUCCUUCG--------------------- (((((((.((...((.(((.((.(((.(((((((((((((......))))...)))))))))))))).)))))...)).))))))).............--------------------- ( -31.90) >DroYak_CAF1 40337 120 - 1 CCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAAUGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCG (((((((.((...(..(((.((.(((.(((((((((((((......))))...)))))))))))))).)))..)..)).))))))).((.(((.(((((........))))))))))... ( -39.70) >consensus CCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAACGGUCCGGGGAACGGGCGCCGGUUCUUCGUGUCUGAUUGAAGGCCGCUCG (((((((.((...(..(((.((.(((.(((((((((((((......))))...)))))))))))))).)))..)..)).)))))))...((((.(((((........))))))))).... (-32.84 = -34.52 + 1.68)


| Location | 22,428,580 – 22,428,700 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22428580 120 - 23771897 UCUUGCACUUUUUCAGCUUUAGUCACACGUUAGAACGGGACCCGUUGGUUUUUGUUGUUAGCACAUUGACACGUUGUGCAUAAAUUUGCAUUUAAAUGUGCCAUGGCGAACGGCCCGGGG .......((..((((((...........))).)))..)).((((..(((((((((..(..(((((((........(((((......)))))...))))))).)..))))).)))))))). ( -30.70) >DroSec_CAF1 282 120 - 1 UCUUGCACUUUUUCAGCUGUAGUCACACGUUAGAGCGGCACCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGG .((((..........(((((..((........)))))))..((((((((...(((.....)))(((.(((((((((((((......))))...))))))))))))))))))))..)))). ( -33.30) >DroSim_CAF1 5188 120 - 1 UCUUGCACUUUUUCAGCUUUAGUCACACGUUAGAGCGGCACCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAGCGGUCCGGGG ..........(..(.(((((((.(....))))))))((...((((((((...(((.....)))(((.(((((((((((((......))))...)))))))))))))))))))).)))..) ( -34.10) >DroEre_CAF1 6329 120 - 1 CCCUGCACUUUUUCAGCUUUAGUCACACGUUAGAGCGGCACCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAACGCUGCAGGG (((((((........(((((((.(....))))))))......(((((((...(((.....)))(((.(((((((((((((......))))...))))))))))))))))))).))))))) ( -40.10) >DroYak_CAF1 40377 120 - 1 UCUUGCACUUUUUCAGCUUGGGUCACACGUUAGAGCAGCACCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAAUGGUCCGGGG ................(((((..((..(((((.(((((((............))))))).......((((((((((((((......))))...)))))))))))))))..))..))))). ( -35.00) >consensus UCUUGCACUUUUUCAGCUUUAGUCACACGUUAGAGCGGCACCCGUUUGUUUUUGUUGUUAGCACAUUGACACGUUUUGCAUAAAUUUGCAUUCAAAUGUGUCAUGGCGAACGGUCCGGGG (((((..........(((((((.(....)))))))).....((((((((...(((.....)))(((.(((((((((((((......))))...))))))))))))))))))))..))))) (-28.82 = -29.18 + 0.36)

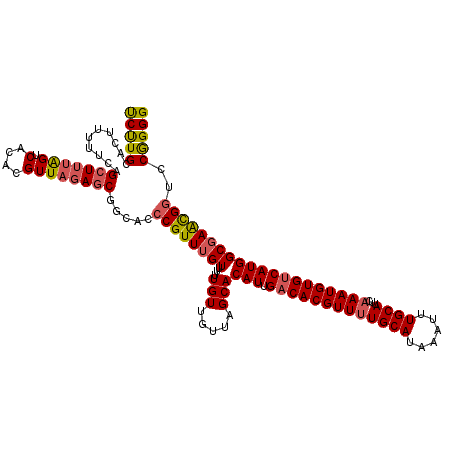

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:35 2006