| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
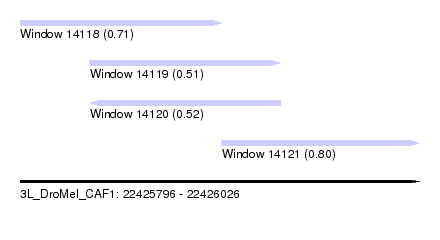
| Location | 22,425,796 – 22,426,026 |
| Length | 230 |
| Max. P | 0.801900 |

| Location | 22,425,796 – 22,425,912 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -28.15 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22425796 116 + 23771897 AAAAAUUAUACUCCAUUUAAUUUUCCAGUUCAGUUCUUGUCUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCCUCCGGGUGUC----UGUUCCCGGAG .(((((((.........)))))))...(((.(((....(.((((((..(((((....)))))..)))))).).....))).)))..........(((((((....----....))))))) ( -31.40) >DroSec_CAF1 2978 120 + 1 AAAAAUUAUACUCCAUUUAAUUUUCCAGUUCAGUUCUUGUCUGGUCCCGGAACGCUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAG .(((((((.........)))))))...(((.(((....(.((((((..(((((....)))))..)))))).).....))).)))............(((((.((......)).))))).. ( -29.50) >DroSim_CAF1 2315 120 + 1 AAAAAUUAUACUCCAUUUAAUUUUCCAGUUCAGUUCUUGUCUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAG .(((((((.........)))))))...(((.(((....(.((((((..(((((....)))))..)))))).).....))).)))............(((((.((......)).))))).. ( -29.50) >consensus AAAAAUUAUACUCCAUUUAAUUUUCCAGUUCAGUUCUUGUCUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAG ..........((((.............(((.(((....(.((((((..(((((....)))))..)))))).).....))).)))..............(((.(........).))))))) (-28.15 = -27.93 + -0.22)



| Location | 22,425,836 – 22,425,946 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -39.13 |
| Energy contribution | -38.47 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22425836 110 + 23771897 CUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCCUCCGGGUGUC----UGUUCCCGGAGCCGGUGUUUGGUGUCAAGUUUGACUGGCCCAGGC (((.....(((((....)))))..((((((((.....(((..(((((.(.(((.(((((((....----....))))))).))).)...)))))..)))..))))))))))).. ( -46.00) >DroSec_CAF1 3018 114 + 1 CUGGUCCCGGAACGCUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAGCCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGC (((.....(((((....)))))..((((((((.....(((..(((((.....(((((((((.((......)).))))).....))))..)))))..)))..))))))))))).. ( -41.50) >DroSim_CAF1 2355 114 + 1 CUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAGCCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGC (((.....(((((....)))))..((((((((.....(((..(((((.....(((((((((.((......)).))))).....))))..)))))..)))..))))))))))).. ( -41.50) >consensus CUGGUCCCGGAACGGUCGUUUCCCGGCCAGUCCUUCUGCUCAACAUCUCCCUGCACCCGGGUGUCUAUCUGCUCCCGGAGCCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGC (((.....(((((....)))))..((((((((.....(((..(((((.....(((.(((((.(........).)))))....)))....)))))..)))..))))))))))).. (-39.13 = -38.47 + -0.66)


| Location | 22,425,836 – 22,425,946 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -40.92 |
| Energy contribution | -40.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22425836 110 - 23771897 GCCUGGGCCAGUCAAACUUGACACCAAACACCGGCUCCGGGAACA----GACACCCGGAGGCAGGGAGAUGUUGAGCAGAAGGACUGGCCGGGAAACGACCGUUCCGGGACCAG .((((((((((((...((..(((.(.....(((.(((((((....----....))))))).).))..).)))..))......)))))))(((.......)))..)))))..... ( -46.70) >DroSec_CAF1 3018 114 - 1 GCCUGGGCCAGUCAAACUUGACACCAAACACAGGCUCCGGGAGCAGAUAGACACCCGGGUGCAGGGAGAUGUUGAGCAGAAGGACUGGCCGGGAAACGAGCGUUCCGGGACCAG .((((((((((((...((..(((.(.....(..((((((((.(........).)))))).))..)..).)))..))......)))))))).(....)........))))..... ( -41.20) >DroSim_CAF1 2355 114 - 1 GCCUGGGCCAGUCAAACUUGACACCAAACACAGGCUCCGGGAGCAGAUAGACACCCGGGUGCAGGGAGAUGUUGAGCAGAAGGACUGGCCGGGAAACGACCGUUCCGGGACCAG .((((((((((((...((..(((.(.....(..((((((((.(........).)))))).))..)..).)))..))......)))))))(((.......)))..)))))..... ( -41.70) >consensus GCCUGGGCCAGUCAAACUUGACACCAAACACAGGCUCCGGGAGCAGAUAGACACCCGGGUGCAGGGAGAUGUUGAGCAGAAGGACUGGCCGGGAAACGACCGUUCCGGGACCAG .((((((((((((...((..(((.(...(....((((((((............)))))).))..)..).)))..))......)))))))).(....)........))))..... (-40.92 = -40.70 + -0.22)



| Location | 22,425,912 – 22,426,026 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 98.25 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -34.43 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22425912 114 + 23771897 CCGGUGUUUGGUGUCAAGUUUGACUGGCCCAGGCUCGGCGCCACUACGGGUUUUAGGUUUUGAGCUCGGGUUCAGGUUUCUGAGUUGACGUUAUGCAUUGAAAUUAAUUAAAUC .((((((.(..((((((.((.((((.((((..((((((.(((.(....)......))).))))))..))))...))))...)).))))))..).)))))).............. ( -38.60) >DroSec_CAF1 3098 114 + 1 CCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGCUCGGCGGCACUACGGGUUUUAGGUUUUGAGCUUGGGUUCAGGUUUCUGAGUUGACGUUAUGCAUUGAAAUUAAUUAAAUC .(.((((.(..((((((.((.((((.((((((((((((.(((.(((.......))))))))))))))))))...))))...)).))))))..).)))).).............. ( -37.00) >DroSim_CAF1 2435 114 + 1 CCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGCUCGGCGGCACUACGGGUUUUAGGUUUUGAGCUUGGGUUCAGGUUUCUGAGUUGACGUUAUGCAUUGAAAUUAAUUAAAUC .(.((((.(..((((((.((.((((.((((((((((((.(((.(((.......))))))))))))))))))...))))...)).))))))..).)))).).............. ( -37.00) >consensus CCUGUGUUUGGUGUCAAGUUUGACUGGCCCAGGCUCGGCGGCACUACGGGUUUUAGGUUUUGAGCUUGGGUUCAGGUUUCUGAGUUGACGUUAUGCAUUGAAAUUAAUUAAAUC .(.((((.(..((((((.((.((((.((((((((((((.(((.(((.......))))))))))))))))))...))))...)).))))))..).)))).).............. (-34.43 = -35.10 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:30 2006