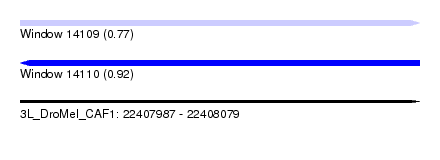
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,407,987 – 22,408,079 |
| Length | 92 |
| Max. P | 0.923942 |

| Location | 22,407,987 – 22,408,079 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.17 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.51 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22407987 92 + 23771897 GAUUUUGUGCUGCGUCAGAGGUUACUUGGGCGCAGCACAGUAACUUGGGAGGUUCAAGA---G---------GUCAGCUUCUCCCGAAAUC------CUCUACAACGAUC ((((((((((((((((.(((....))).))))))))))))....(((((((((((.(((---(---------(....)))))...)))..)------)))).))).)))) ( -35.00) >DroGri_CAF1 8812 102 + 1 UAUUUUGUGCUGCGUCAUACUC-------AAGUGGCACAAAAAAGAG-UAAGCUGAAUUUAUGUUCAUAUUUGUUGUUGUUGGCGAAGCUUUUGUAUUUUUUGUAUGAUC ...(((((((..(.........-------..)..)))))))((((((-((((((((((....)))))..((((((......)))))))))....))))))))........ ( -22.00) >DroSec_CAF1 7365 85 + 1 GAUUUUGUGCUGCGUCAGAGUU-------GCGCAGCACUGUAACUUGGGAGGUUCAAGA---G---------GUCAGCUUCUCCCGAAAUC------CUCUACAACGAUC (((((((((((((((.......-------)))))))))........((((((..(....---.---------....)..))))))))))))------............. ( -27.40) >DroSim_CAF1 7381 85 + 1 GAUUUUGUGCUGCGUCAGAGUU-------GCGCAGCACUGUAACUUGGGAGGUUCAAGA---G---------GUCAGCUUCUCCCGAAAUC------CUCUACAACGAUC (((((((((((((((.......-------)))))))))........((((((..(....---.---------....)..))))))))))))------............. ( -27.40) >DroEre_CAF1 7654 78 + 1 GAUUUUGUGCUGCGUCAGAGGU-------GCGCGGCACU------ACGUAAGUCCAAAA---G---------GUCAGCUUCUACCGCAAUG------CUCCACAACGUU- ......(((((((((.......-------))))))))).------((((..(.((....---)---------).).((.......))....------.......)))).- ( -19.90) >DroYak_CAF1 7101 83 + 1 GAUUUUGUGCUGCGUCAGAGGU-------GCGCAGCACUAUA--UACAUAAGUCGAAGA---G---------GUCAGCUUCUGCCGAAAUA------CUCUACAACGAUC ......(((((((((.......-------)))))))))....--.......((((.(((---(---------.((.((....)).))....------))))....)))). ( -24.20) >consensus GAUUUUGUGCUGCGUCAGAGGU_______GCGCAGCACUGUAACUAGGGAAGUUCAAGA___G_________GUCAGCUUCUCCCGAAAUC______CUCUACAACGAUC ......(((((((((..............)))))))))........................................................................ (-13.84 = -13.51 + -0.33)


| Location | 22,407,987 – 22,408,079 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.17 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22407987 92 - 23771897 GAUCGUUGUAGAG------GAUUUCGGGAGAAGCUGAC---------C---UCUUGAACCUCCCAAGUUACUGUGCUGCGCCCAAGUAACCUCUGACGCAGCACAAAAUC ..........(((------(..((((((((........---------)---))))))))))).........(((((((((....((......))..)))))))))..... ( -29.00) >DroGri_CAF1 8812 102 - 1 GAUCAUACAAAAAAUACAAAAGCUUCGCCAACAACAACAAAUAUGAACAUAAAUUCAGCUUA-CUCUUUUUUGUGCCACUU-------GAGUAUGACGCAGCACAAAAUA .....................(((.((.((.............((((......))))...((-(((...............-------))))))).)).)))........ ( -10.46) >DroSec_CAF1 7365 85 - 1 GAUCGUUGUAGAG------GAUUUCGGGAGAAGCUGAC---------C---UCUUGAACCUCCCAAGUUACAGUGCUGCGC-------AACUCUGACGCAGCACAAAAUC ......(((((.(------((.((((((((........---------)---)))))))..)))....)))))(((((((((-------(....)).))))))))...... ( -29.70) >DroSim_CAF1 7381 85 - 1 GAUCGUUGUAGAG------GAUUUCGGGAGAAGCUGAC---------C---UCUUGAACCUCCCAAGUUACAGUGCUGCGC-------AACUCUGACGCAGCACAAAAUC ......(((((.(------((.((((((((........---------)---)))))))..)))....)))))(((((((((-------(....)).))))))))...... ( -29.70) >DroEre_CAF1 7654 78 - 1 -AACGUUGUGGAG------CAUUGCGGUAGAAGCUGAC---------C---UUUUGGACUUACGU------AGUGCCGCGC-------ACCUCUGACGCAGCACAAAAUC -....((((((.(------(((((((....((((..(.---------.---..)..).))).)))------))))))((((-------(....)).)))..))))).... ( -22.30) >DroYak_CAF1 7101 83 - 1 GAUCGUUGUAGAG------UAUUUCGGCAGAAGCUGAC---------C---UCUUCGACUUAUGUA--UAUAGUGCUGCGC-------ACCUCUGACGCAGCACAAAAUC ....((((.((((------....(((((....))))).---------)---))).)))).......--....(((((((((-------(....)).))))))))...... ( -29.00) >consensus GAUCGUUGUAGAG______GAUUUCGGGAGAAGCUGAC_________C___UCUUGAACCUACCAAGUUACAGUGCUGCGC_______AACUCUGACGCAGCACAAAAUC .......................((((......))))...................................((((((((................))))))))...... (-10.98 = -11.87 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:20 2006