
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,387,707 – 22,387,815 |
| Length | 108 |
| Max. P | 0.972611 |

| Location | 22,387,707 – 22,387,815 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -25.39 |
| Energy contribution | -26.70 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
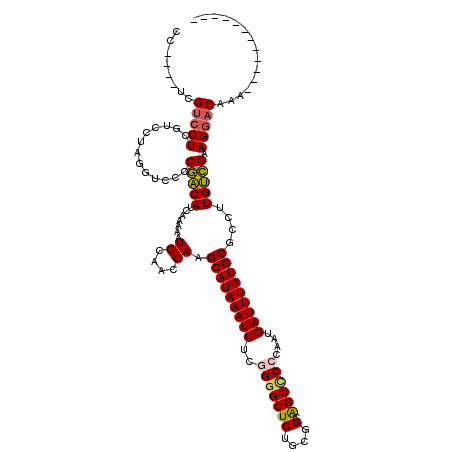
>3L_DroMel_CAF1 22387707 108 + 23771897 ------------UUUGUCCUUGGACAAGGCGCAUAAAUCAAUGGGGGCCGCCGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGUCCGGGUCCUAGCACGAGGACGAGGAUGG ------------((((((((((..(.((((((((((((...(.((((((......).))))).).))))))))((((..(((((...)))))))))).))).)..))))))))))..... ( -40.00) >DroPse_CAF1 93428 107 + 1 UGCUGGCGACGUUUUGUCCUUGAGCAGGACGCAUAAAUCAUA--GAGCUGCUGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGCUCC--ACCCACGA--AGGACG------- ...............((((((..((((.(.((..........--..))).)))).((((..(((((...((((......)))))))))..)))).--.......)--))))).------- ( -26.90) >DroSec_CAF1 38830 108 + 1 ------------UUUGUCCUUGGACAAGGCGCAUAAAUCAUCGGGGGCUGCCGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGUCCGGGACCUAGGACGAGGACGAGGAUGG ------------((((((((((..(.((((((((((((..((((((.(((...)))..)))))).))))))))((((..(((((...)))))))))).))).)..))))))))))..... ( -44.10) >DroEre_CAF1 55117 105 + 1 ------------UUUGUCCUUGGACAAGGCGCAUAAAUCAUUGGGGGCUGCCGCAGAGCUCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGUCCGGGACUUAGGACGAGCCCGU---CCU ------------...(((((.(((((....((((((((..((.((((((.......)))))).)))))))))).......((....)).))))))))))..((((((....)))---))) ( -39.30) >DroYak_CAF1 55350 108 + 1 ------------UUUGUCCUUGGACAAGGCGCAUAAAUCAUUGGGGGCUGCCGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGUCCGGGACCUAGGAUGAGGACGAGGACGG ------------((((((((((..(.((((((((((((..((.((((((.......)))))).))))))))))((((..(((((...)))))))))).))).)..))))))))))..... ( -40.40) >DroPer_CAF1 96030 107 + 1 UGCUGGCGACGUUUUGUCCUUGAGCAGGACGCAUAAAUCAUA--GAGCUGCUGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGCUCC--ACCCACGA--AGGACG------- ...............((((((..((((.(.((..........--..))).)))).((((..(((((...((((......)))))))))..)))).--.......)--))))).------- ( -26.90) >consensus ____________UUUGUCCUUGGACAAGGCGCAUAAAUCAUUGGGGGCUGCCGCAGAGCCCCGAAAUUUAUGCUUGGUUGCAUUUUUGAUGUCCGGGACCUAGGACGAGGACGA____GG ...............(((((((..(.(((.((((((((.....((((((.......))))))...))))))))..((..(((((...)))))))....))).)..)))))))........ (-25.39 = -26.70 + 1.31)



| Location | 22,387,707 – 22,387,815 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -26.07 |
| Energy contribution | -25.71 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22387707 108 - 23771897 CCAUCCUCGUCCUCGUGCUAGGACCCGGACAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCGGCGGCCCCCAUUGAUUUAUGCGCCUUGUCCAAGGACAAA------------ ...((((.(((((......)))))..(((((.......((....)).(((((((((..((((((.......))))))....)))))))))....))))).))))....------------ ( -35.20) >DroPse_CAF1 93428 107 - 1 -------CGUCCU--UCGUGGGU--GGAGCAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCAGCAGCUC--UAUGAUUUAUGCGUCCUGCUCAAGGACAAAACGUCGCCAGCA -------.(((((--(.(..(((--((.((((.....))))..))).(((((((((..(((((((....).)))))--)..)))))))))..))..)..))))))............... ( -36.60) >DroSec_CAF1 38830 108 - 1 CCAUCCUCGUCCUCGUCCUAGGUCCCGGACAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCGGCAGCCCCCGAUGAUUUAUGCGCCUUGUCCAAGGACAAA------------ ........(((((.(..(.((((...((.(((.....)))...))..((((((((((((((((((....).)).)))))).))))))))))))).)..).)))))...------------ ( -36.40) >DroEre_CAF1 55117 105 - 1 AGG---ACGGGCUCGUCCUAAGUCCCGGACAUCAAAAAUGCAACCAAGCAUAAAUUUCGGAGCUCUGCGGCAGCCCCCAAUGAUUUAUGCGCCUUGUCCAAGGACAAA------------ (((---(((....))))))..((((.(((((.......((....)).(((((((((..((.((((....).))).))....)))))))))....)))))..))))...------------ ( -33.50) >DroYak_CAF1 55350 108 - 1 CCGUCCUCGUCCUCAUCCUAGGUCCCGGACAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCGGCAGCCCCCAAUGAUUUAUGCGCCUUGUCCAAGGACAAA------------ ..(((((.(.(((......))).)..(((((.......((....)).(((((((((..(((((((....).))))))....)))))))))....))))).)))))...------------ ( -35.80) >DroPer_CAF1 96030 107 - 1 -------CGUCCU--UCGUGGGU--GGAGCAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCAGCAGCUC--UAUGAUUUAUGCGUCCUGCUCAAGGACAAAACGUCGCCAGCA -------.(((((--(.(..(((--((.((((.....))))..))).(((((((((..(((((((....).)))))--)..)))))))))..))..)..))))))............... ( -36.60) >consensus CC____UCGUCCUCGUCCUAGGUCCCGGACAUCAAAAAUGCAACCAAGCAUAAAUUUCGGGGCUCUGCGGCAGCCCCCAAUGAUUUAUGCGCCUUGUCCAAGGACAAA____________ ........(((((.............(((((.......((....)).(((((((((..(((((((....).))))))....)))))))))....))))).)))))............... (-26.07 = -25.71 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:20:08 2006