
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,386,136 – 22,386,315 |
| Length | 179 |
| Max. P | 0.987493 |

| Location | 22,386,136 – 22,386,239 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.40 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22386136 103 + 23771897 AAAUAAGUCAAC--------ACUGG------CGAGUGGAUGUAUAUAUAUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ......(((...--------.(((.------((((..(((((((....)))))))..---))((((....)))).)).)))............(((.(((((....))))).))).))). ( -29.50) >DroPse_CAF1 91503 118 + 1 AAAUAAGUCAGCCAGCACACACACACGUACACAGAAGAAUGUCGG--UGUAUGUCUGGCACUGGGAGACUCCUCCUAGUGGACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ......(((.(((((.(((.((((.((.(((........))))))--))).))))))))((((((((....)))))))).)))..........(((.(((((....))))).)))..... ( -43.50) >DroSim_CAF1 52847 99 + 1 AAAUAAGUCAAC--------ACUGG------CGAGAGGAUGUA----UGUGCGUCUG---CUGGAAAAGUUUCCCCGGCGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ............--------...((------(...........----((((.(((((---((((..........)))))))))))))......(((.(((((....))))).)))..))) ( -33.00) >DroEre_CAF1 53512 99 + 1 AAAUAAGUCAAC--------ACUGG------CGAGAGGAUGUG----UGUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ............--------...((------(.......((((----.((.((((..---..((((....))))..)))).))))))......(((.(((((....))))).)))..))) ( -28.20) >DroYak_CAF1 53802 99 + 1 AAAUAAGUCAAC--------ACUGG------CUACAGGAUGUA----UGUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ......(((...--------.(((.------...)))......----((((.(((((---(.((((....))))..).)))))))))......(((.(((((....))))).))).))). ( -27.40) >DroPer_CAF1 94104 118 + 1 AAAUAAGUCAGCCAGCACACACACACGUACACAGAAGAAUGUCGG--UGUAUGUCUGGCACUGGGAGACUCCUCCUAGUGGACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ......(((.(((((.(((.((((.((.(((........))))))--))).))))))))((((((((....)))))))).)))..........(((.(((((....))))).)))..... ( -43.50) >consensus AAAUAAGUCAAC________ACUGG______CGAGAGGAUGUA____UGUGCGUCUG___CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCC ......(((......................................((((.(((((.....(((......)))....)))))))))......(((.(((((....))))).))).))). (-19.45 = -19.40 + -0.05)



| Location | 22,386,136 – 22,386,239 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -16.55 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22386136 103 - 23771897 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCAUAUAUAUACAUCCACUCG------CCAGU--------GUUGACUUAUUU (((..(((.(((((....))))).)))......(((((((.((.((..(....)..)).)---).))).))))...............)------))...--------............ ( -25.70) >DroPse_CAF1 91503 118 - 1 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGGGGUCCACUAGGAGGAGUCUCCCAGUGCCAGACAUACA--CCGACAUUCUUCUGUGUACGUGUGUGUGUGCUGGCUGACUUAUUU ((((((((.(((((....))))).)))........))))).(((.((((....)))).)))((((((((((((--(((((((......)))).)).)))))))).))))).......... ( -46.20) >DroSim_CAF1 52847 99 - 1 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGCCGGGGAAACUUUUCCAG---CAGACGCACA----UACAUCCUCUCG------CCAGU--------GUUGACUUAUUU (((..(((.(((((....))))).)))......(((((((.((.((..(....)..)).)---).))).))))----...........)------))...--------............ ( -29.20) >DroEre_CAF1 53512 99 - 1 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCACA----CACAUCCUCUCG------CCAGU--------GUUGACUUAUUU (((..(((.(((((....))))).)))......((((((.((((..((((....))))..---..)))).)).----)))).......)------))...--------............ ( -29.10) >DroYak_CAF1 53802 99 - 1 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCACA----UACAUCCUGUAG------CCAGU--------GUUGACUUAUUU (((..(((.(((((....))))).)))......(((((((.((.((..(....)..)).)---).))).))))----...........)------))...--------............ ( -27.50) >DroPer_CAF1 94104 118 - 1 GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGGGGUCCACUAGGAGGAGUCUCCCAGUGCCAGACAUACA--CCGACAUUCUUCUGUGUACGUGUGUGUGUGCUGGCUGACUUAUUU ((((((((.(((((....))))).)))........))))).(((.((((....)))).)))((((((((((((--(((((((......)))).)).)))))))).))))).......... ( -46.20) >consensus GGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGCCGGGGAAACUUUUCCAG___CAGACGCACA____UACAUCCUCUCG______CCAGU________GUUGACUUAUUU (((..(((.(((((....))))).)))......(((((((....(((((....))))).......))).))))...................................)))......... (-16.55 = -15.75 + -0.80)

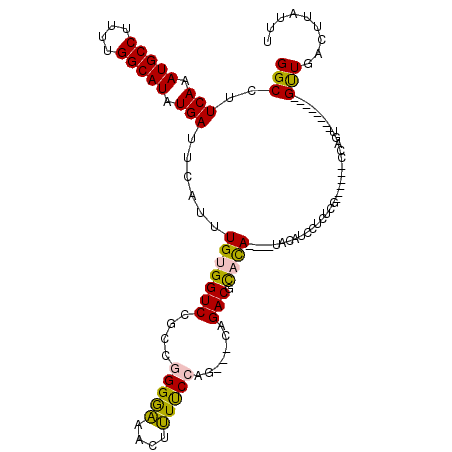
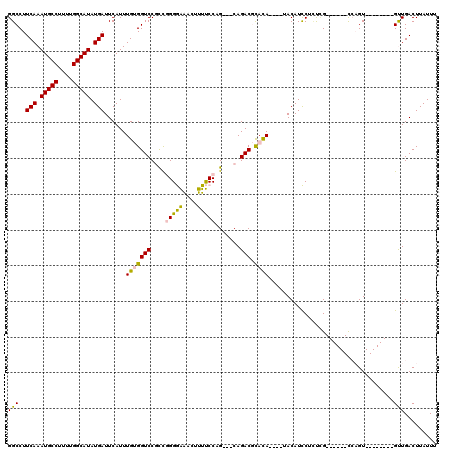
| Location | 22,386,162 – 22,386,279 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -27.56 |
| Energy contribution | -26.62 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22386162 117 + 23771897 GUAUAUAUAUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGGCCUUUGGGUUCUUCUCCAGCC ((((....))))....(---(((((.(((....(((((...............(((.(((((....))))).)))((((((((.((....))..)))))))))))))..))).)))))). ( -40.10) >DroPse_CAF1 91543 114 + 1 GUCGG--UGUAUGUCUGGCACUGGGAGACUCCUCCUAGUGGACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCUUUUGUAAUGGGCCCUUUGGGUCCAACUCC---- (((((--.......)))))((((((((....))))))))((((((........(((.(((((....))))).))).(((((...((....))...)))))....))))))......---- ( -46.80) >DroSim_CAF1 52873 113 + 1 GUA----UGUGCGUCUG---CUGGAAAAGUUUCCCCGGCGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGCCCUUUGGGUCCUUCGCCAGCC ...----((((.(((((---((((..........)))))))))))))...((((((.(((((....))))).)))((((.....)))).......((((((...)))))))))....... ( -42.90) >DroEre_CAF1 53538 113 + 1 GUG----UGUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCAUUGUAAUGGGGCCUUUGGGUCCUCCUCCAGCC ...----.........(---(((((...(((.....)))(((((.((((....(((.(((((....))))).))).(((((((.((....))..))))))))))))))))...)))))). ( -41.80) >DroYak_CAF1 53828 104 + 1 GUA----UGUGCGUCUG---CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCCUAAGCCAUUGUAAUGGGGCCUUUGGGUCCU--------- ...----.....(((..---..((((....))))..)))(((((.((((....(((.(((((....))))).))).(((((((.((....))..)))))))))))))))).--------- ( -37.60) >DroPer_CAF1 94144 114 + 1 GUCGG--UGUAUGUCUGGCACUGGGAGACUCCUCCUAGUGGACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCUUUUGUAAUGGGCCCUUUGGGUCCAACUCC---- (((((--.......)))))((((((((....))))))))((((((........(((.(((((....))))).))).(((((...((....))...)))))....))))))......---- ( -46.80) >consensus GUA____UGUGCGUCUG___CUGGAAAAGUUUCCCCGACGGACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGCCCUUUGGGUCCUACUCC____ ......................(((......))).....(((((.((((....(((.(((((....))))).))).(((((...((....))...))).))))))))))).......... (-27.56 = -26.62 + -0.94)


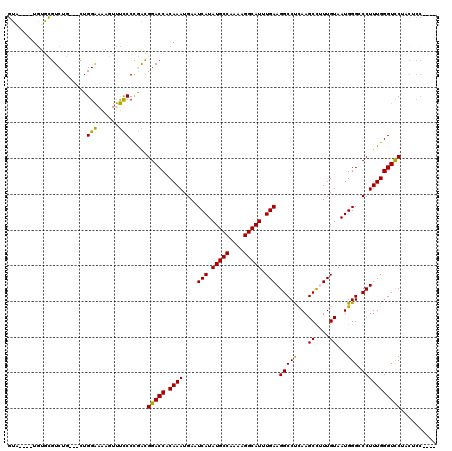
| Location | 22,386,162 – 22,386,279 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -42.53 |
| Consensus MFE | -27.07 |
| Energy contribution | -26.85 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22386162 117 - 23771897 GGCUGGAGAAGAACCCAAAGGCCCCAUUACAAAGGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCAUAUAUAUAC .((((((((((..(((...(((...(((((((((((.....))).(((.(((((....))))).)))....))))))))..))).)))...)))))))))---)................ ( -43.40) >DroPse_CAF1 91543 114 - 1 ----GGAGUUGGACCCAAAGGGCCCAUUACAAAAGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGGGGUCCACUAGGAGGAGUCUCCCAGUGCCAGACAUACA--CCGAC ----...(((((.(((((((((((((...........)).)))))(((.(((((....))))).)))....))))))(((((((.((((....)))).))))...))).....--))))) ( -43.00) >DroSim_CAF1 52873 113 - 1 GGCUGGCGAAGGACCCAAAGGGCCCAUUACAAAGGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGCCGGGGAAACUUUUCCAG---CAGACGCACA----UAC .(((((.((((..(((...(((((....((((((((.....))).(((.(((((....))))).)))....))))))))))....)))...)))).))))---).........----... ( -43.00) >DroEre_CAF1 53538 113 - 1 GGCUGGAGGAGGACCCAAAGGCCCCAUUACAAUGGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCACA----CAC .(((((((..(((((((((((((((((....)))).....)))).(((.(((((....))))).)))....)))).)))))..)(((....))).)))))---).........----... ( -45.00) >DroYak_CAF1 53828 104 - 1 ---------AGGACCCAAAGGCCCCAUUACAAUGGCUUAGGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGUCGGGGAAACUUUUCCAG---CAGACGCACA----UAC ---------.((((((((((((((...............))))).(((.(((((....))))).)))....)))).)))))(((..((((....))))..---..))).....----... ( -37.76) >DroPer_CAF1 94144 114 - 1 ----GGAGUUGGACCCAAAGGGCCCAUUACAAAAGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGGGGUCCACUAGGAGGAGUCUCCCAGUGCCAGACAUACA--CCGAC ----...(((((.(((((((((((((...........)).)))))(((.(((((....))))).)))....))))))(((((((.((((....)))).))))...))).....--))))) ( -43.00) >consensus ____GGAGAAGGACCCAAAGGCCCCAUUACAAAGGCUUGAGGCCUUCAAAUGCCUUUUGGCAUAUGAUUCAUUUGUGGUCCGCCGGGGAAACUUUUCCAG___CAGACGCACA____UAC ..........(((((((((((.((((...........)).)))).(((.(((((....))))).)))....)))).)))))...(((((....)))))...................... (-27.07 = -26.85 + -0.22)



| Location | 22,386,199 – 22,386,315 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -22.23 |
| Energy contribution | -21.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22386199 116 + 23771897 GACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGGCCUUUGGGUUCUUCUCCAGCCCACC-UCCAU---CCUCCUCGACAUGUCUGCGCCAACAUG .............(((.(((((....))))).)))((((((((.((....))..)))))))).((((((.......))))))..-.....---.........(((((........))))) ( -29.70) >DroPse_CAF1 91581 111 + 1 GACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCUUUUGUAAUGGGCCCUUUGGGUCCAACUCC---------UUCAUUUCACUCCUGGUUUUGUCUGCCUCAAAAUG (((((........(((.(((((....))))).))).(((((...((....))...)))))....))))).......---------...............((((((.......)))))). ( -28.60) >DroSim_CAF1 52906 116 + 1 GACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGCCCUUUGGGUCCUUCGCCAGCCCACC-UCCAU---CCUCCUCGCCAUGUCUGCGCCAACAUG ....(((((....(((.(((((....))))).))).(((.....))))))))..(((((....(((........))))))))..-.....---.........(((((........))))) ( -27.00) >DroEre_CAF1 53571 117 + 1 GACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCAUUGUAAUGGGGCCUUUGGGUCCUCCUCCAGCCCAUCAUCCGU---CCUCCUCGCCAUGUCAGCGCCAACAUG (((.....((((.(((.(((((....))))).)))((((((((.((....))..)))))))).(((((.........)))))))))..))---)........(((((........))))) ( -30.80) >DroYak_CAF1 53861 95 + 1 GACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCCUAAGCCAUUGUAAUGGGGCCUUUGGGUCCU-------------------------CCUCGCCAUGUCAGCCCCAACAUG ((((.((((....(((.(((((....))))).))).(((((((.((....))..)))))))))))))))..-------------------------......(((((........))))) ( -31.10) >DroPer_CAF1 94182 111 + 1 GACCCCAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCUUUUGUAAUGGGCCCUUUGGGUCCAACUCC---------UUCAUUUCACUCCUGGUUUUGUCUGCCUCAAAAUG (((((........(((.(((((....))))).))).(((((...((....))...)))))....))))).......---------...............((((((.......)))))). ( -28.60) >consensus GACCACAAAUGAAUCAUAUGCCAAAAGGCAUUUGAAGGCCUCAAGCCUUUGUAAUGGGCCCUUUGGGUCCUACUCC_________UCCAU___CCUCCUCGCCAUGUCUGCCCCAACAUG ((((.((((....(((.(((((....))))).))).(((((...((....))...))).))))))))))..............................(((.......)))........ (-22.23 = -21.78 + -0.44)


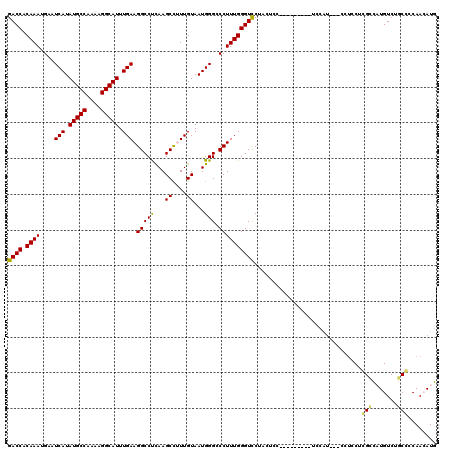
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:59 2006