
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,384,853 – 22,384,973 |
| Length | 120 |
| Max. P | 0.940488 |

| Location | 22,384,853 – 22,384,954 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22384853 101 + 23771897 CAACAGUAACUCUUUCUUGCAGUCUUCAUAAAUUUAAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAAGCCGCGUCUGGGCCA ..................(((........................)))((((((((((..........(((((.(((....)))))))))))).)))))). ( -22.36) >DroSec_CAF1 35970 101 + 1 CGACCGUAACUCUUUCUUGCAGUCUUCAUAAAUUUCAUUUUUUCAUGCGGUCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGCCUGGGCCA .(((((((.........((.(((........))).))........)))))))........(.((((..(((.(((((....))))).....))).))))). ( -23.33) >DroSim_CAF1 51554 101 + 1 CGACCGUAACUCUUUCUUGCAGUCUUCAUAAAUUUCAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGCCUGGGCCA .(((.((((.......)))).)))........................((((((......((((....(((..((((....))))))).)))).)))))). ( -25.70) >DroEre_CAF1 52293 101 + 1 CGACCGUAAUUCUUUCUUGCGGUCUUCAUAAAUUCCGUUUUUACAUGCGGCCCAGUGUCAGUGGCCUUGGCUUUGGGCCUGCCCAACACGUCUCUGGGCCA .((((((((.......))))))))........................(((((((.(...(((((....)).(((((....))))))))..).))))))). ( -39.90) >DroYak_CAF1 52529 101 + 1 CGACCGUAAUUCUUUCUUGCGGUCUUCAUAAAUUUAAUUUUUACAUGUGGCCCAGUGUCAGGGCCCUUGGCUUUGGGCCUUGCCAGAACAUCUCUGGGCCA .((((((((.......)))))))).......................((((((((((.((((((((........)))))))).))........)))))))) ( -40.90) >consensus CGACCGUAACUCUUUCUUGCAGUCUUCAUAAAUUUCAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGUCUGGGCCA .(((.((((.......)))).)))........................(((((((.(((((.....))))).)))))))..((((.........))))... (-21.16 = -20.92 + -0.24)

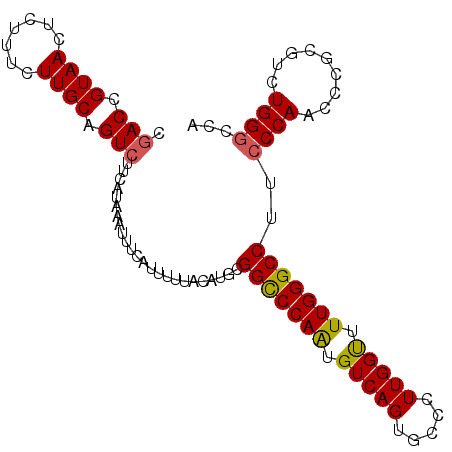
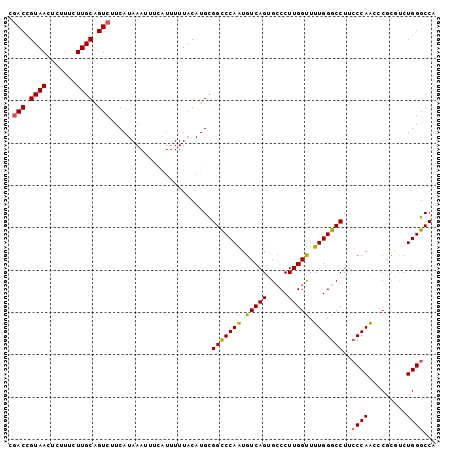
| Location | 22,384,853 – 22,384,954 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22384853 101 - 23771897 UGGCCCAGACGCGGCUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUUAAAUUUAUGAAGACUGCAAGAAAGAGUUACUGUUG .....(((..((((((..(..((((((........)))).))....)..))))))......................((((.(......))))).)))... ( -27.70) >DroSec_CAF1 35970 101 - 1 UGGCCCAGGCGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGACCGCAUGAAAAAAUGAAAUUUAUGAAGACUGCAAGAAAGAGUUACGGUCG .(((((......))))).(..((((((........)))).))..)....(((((((((((.........))))))..((((.(......))))).))))). ( -26.90) >DroSim_CAF1 51554 101 - 1 UGGCCCAGGCGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUGAAAUUUAUGAAGACUGCAAGAAAGAGUUACGGUCG .((((..(((.(((((..(....((((........)))).)..)).))).)))(((.((....((((...))))....))))).............)))). ( -30.20) >DroEre_CAF1 52293 101 - 1 UGGCCCAGAGACGUGUUGGGCAGGCCCAAAGCCAAGGCCACUGACACUGGGCCGCAUGUAAAAACGGAAUUUAUGAAGACCGCAAGAAAGAAUUACGGUCG .(((((((.(.(((((((((....))))).((....))))).).).)))))))........................(((((.((.......)).))))). ( -33.00) >DroYak_CAF1 52529 101 - 1 UGGCCCAGAGAUGUUCUGGCAAGGCCCAAAGCCAAGGGCCCUGACACUGGGCCACAUGUAAAAAUUAAAUUUAUGAAGACCGCAAGAAAGAAUUACGGUCG ((((((((((.....))..((.(((((........))))).))...))))))))((((.............))))..(((((.((.......)).))))). ( -34.62) >consensus UGGCCCAGACGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUGAAAUUUAUGAAGACUGCAAGAAAGAGUUACGGUCG .(((((((......(((((....((((........)))).))))).)))))))........................(((((.((.......)).))))). (-23.26 = -23.90 + 0.64)

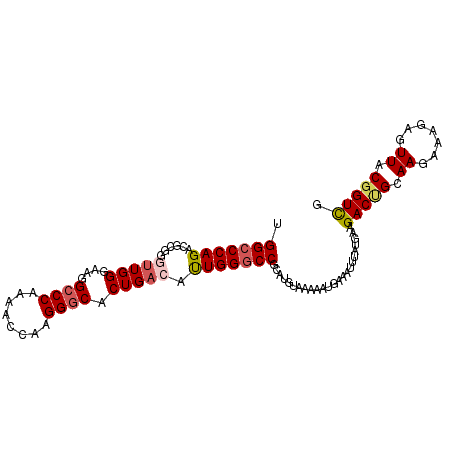
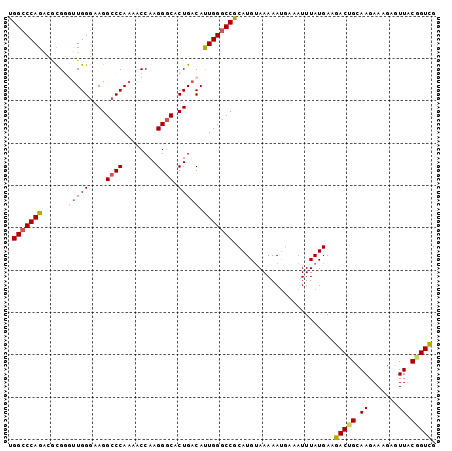
| Location | 22,384,883 – 22,384,973 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -20.66 |
| Energy contribution | -19.78 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22384883 90 + 23771897 AAUUUAAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAAGCCGCGUCUGGGCCAACUGGACCAACCAAAUAUG ................(((((.....(..((.((((........))))))..)...)))))(((..(.....)..)))............ ( -24.70) >DroSec_CAF1 36000 90 + 1 AAUUUCAUUUUUUCAUGCGGUCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGCCUGGGCCAACUGGACCAACCAAAUAUG ..................((((((..((..(.((((..(((.(((((....))))).....))).))))).))))))))........... ( -24.50) >DroSim_CAF1 51584 90 + 1 AAUUUCAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGCCUGGGCCAACUGGACCAACCAAAUAUG .............((((.((((((......((((....(((..((((....))))))).)))).))))))...(((.....)))..)))) ( -23.00) >DroEre_CAF1 52323 90 + 1 AAUUCCGUUUUUACAUGCGGCCCAGUGUCAGUGGCCUUGGCUUUGGGCCUGCCCAACACGUCUCUGGGCCAACUGGACCGUCCAAAUAUG ...(((((........))(((((((.(...(((((....)).(((((....))))))))..).)))))))....)))............. ( -32.00) >DroYak_CAF1 52559 90 + 1 AAUUUAAUUUUUACAUGUGGCCCAGUGUCAGGGCCCUUGGCUUUGGGCCUUGCCAGAACAUCUCUGGGCCAACUGGACCGUCCAAAUAUG .............((((((((((((((.((((((((........)))))))).))........)))))))...(((.....))).))))) ( -34.50) >consensus AAUUUCAUUUUUACAUGCGGCCCAAUGUCAGUGCCCUUGGUUUUGGGCCUUCCCAACCCGCGUCUGGGCCAACUGGACCAACCAAAUAUG .............((((.(((((((.(((((.....))))).)))))))...............(((.((....)).)))......)))) (-20.66 = -19.78 + -0.88)

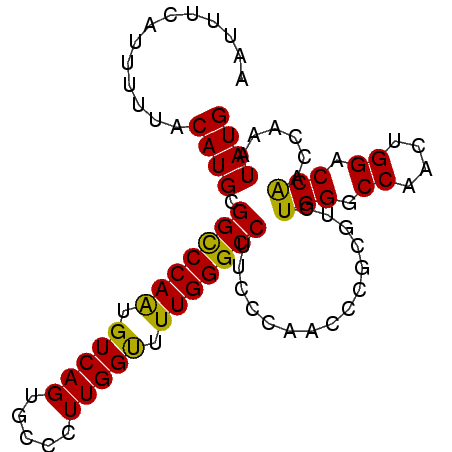
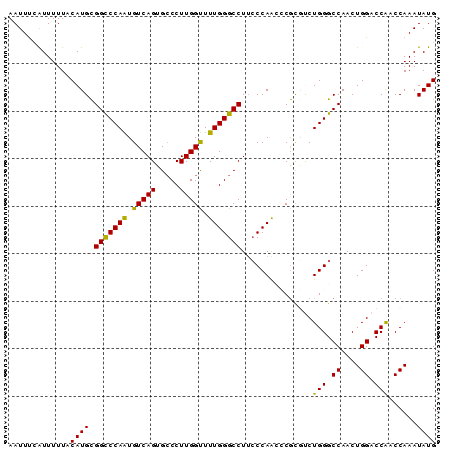
| Location | 22,384,883 – 22,384,973 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22384883 90 - 23771897 CAUAUUUGGUUGGUCCAGUUGGCCCAGACGCGGCUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUUAAAUU ...((((((((((.((....)).))....((((((..(..((((((........)))).))....)..)))))).......)))))))). ( -31.30) >DroSec_CAF1 36000 90 - 1 CAUAUUUGGUUGGUCCAGUUGGCCCAGGCGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGACCGCAUGAAAAAAUGAAAUU ....(((.(((((((((((.(((((......))))).(..((((((........)))).))..)))))))))).)).))).......... ( -29.90) >DroSim_CAF1 51584 90 - 1 CAUAUUUGGUUGGUCCAGUUGGCCCAGGCGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUGAAAUU (((.(((((((((((((((.(((((......))))).(..((((((........)))).))..)))))))))).)).)))).)))..... ( -28.60) >DroEre_CAF1 52323 90 - 1 CAUAUUUGGACGGUCCAGUUGGCCCAGAGACGUGUUGGGCAGGCCCAAAGCCAAGGCCACUGACACUGGGCCGCAUGUAAAAACGGAAUU ..(((...(.(((((((((.(((((((.......)))))).((((.........)))).....)))))))))))..)))........... ( -31.90) >DroYak_CAF1 52559 90 - 1 CAUAUUUGGACGGUCCAGUUGGCCCAGAGAUGUUCUGGCAAGGCCCAAAGCCAAGGGCCCUGACACUGGGCCACAUGUAAAAAUUAAAUU ..(((.((...((((((((.(..((((((...))))))((.(((((........))))).)).))))))))).)).)))........... ( -31.10) >consensus CAUAUUUGGUUGGUCCAGUUGGCCCAGACGCGGGUUGGGAAGGCCCAAAACCAAGGGCACUGACAUUGGGCCGCAUGUAAAAAUGAAAUU ...........((((((((.(.(((((.......))))).((((((........)))).))..))))))))).................. (-24.42 = -24.26 + -0.16)

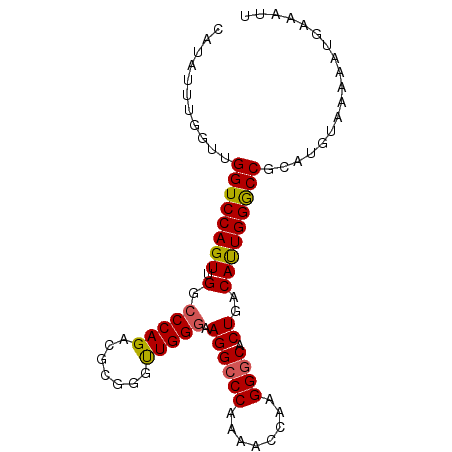
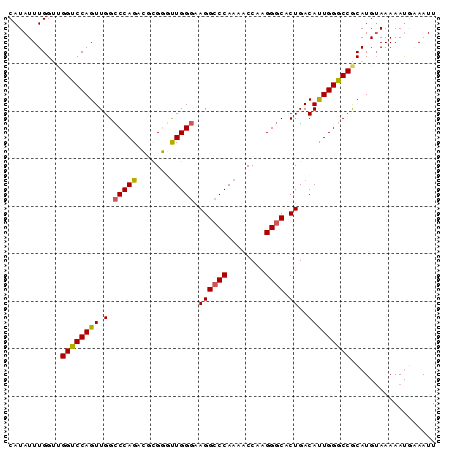
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:55 2006