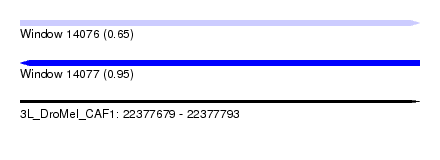
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,377,679 – 22,377,793 |
| Length | 114 |
| Max. P | 0.951886 |

| Location | 22,377,679 – 22,377,793 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -20.89 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
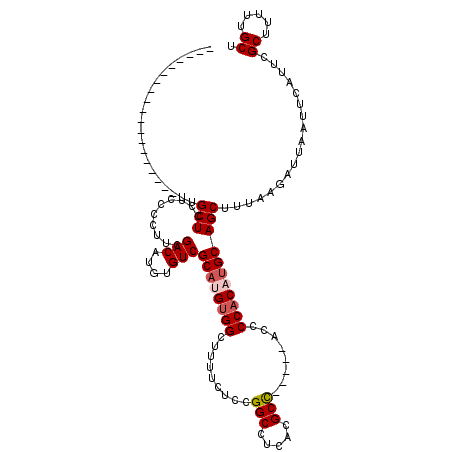
>3L_DroMel_CAF1 22377679 114 + 23771897 AGCAAAAAGCGAAUGAAUUAAUCUUGAUGCUGCAUGUGGGGU-----GGCGUGAGGCCGGAGAAAAGCCACAUGCGACACAUGUCGAAGGGGAGGAGCAAAACAACACACCCCCCCCCC .(((...((((...((.....))....))))...)))((((.-----.(((((.(((.........))).)))))(((....)))...((((.((..............)))))))))) ( -31.54) >DroSec_CAF1 28756 100 + 1 AGCAAAAAGCGAAUGAAUUAAUCUUAAAGCUGCAUGUGGGGUGGAGUGGCGUGAGGCCGGAGAAAAGCCACAUGCGACACAUGUCGAAGGGGAGGAGCAA------------------- .((.....))..................(((((((((((..(....((((.....)))).....)..))))))))(((....)))..........)))..------------------- ( -25.80) >DroSim_CAF1 44299 100 + 1 AGCAAAAAGCGAAUGAAUUAAUCUUAAAGCUGCAUGUGGGGUGGAGUGGCGUGAGGCCGGAGAAAAGCCACAUGCGACACAUGUCGAAGGGGAGGAGCAA------------------- .((.....))..................(((((((((((..(....((((.....)))).....)..))))))))(((....)))..........)))..------------------- ( -25.80) >DroEre_CAF1 45168 93 + 1 AGCAAAAAGCAAAUGAAUUAAUCUUAAAGCUGCAUGUGGGCU-----GGCGGGAGGCCGGAGAAAGGCCACUUGCGACACAUGUCGCAGGGGAGGAGC--------------------- .((.....))...........((((...((.(((((((....-----.(((((.((((.......)))).)))))..))))))).))..)))).....--------------------- ( -30.00) >DroYak_CAF1 38599 95 + 1 AGCAAAAAGCGAAUGAAUUAAUCUUAAAGCUGCAUGUGGGUU-----AGCGUGAGGCCGGAGGAAAGCCGCUAGCGACAGAUGUCGCAGGGGAGGAGCAA------------------- ........((...........((((...(((.(((((.....-----.))))).)))...))))...((.((.(((((....))))))).))....))..------------------- ( -27.20) >consensus AGCAAAAAGCGAAUGAAUUAAUCUUAAAGCUGCAUGUGGGGU_____GGCGUGAGGCCGGAGAAAAGCCACAUGCGACACAUGUCGAAGGGGAGGAGCAA___________________ .((.....))..................(((((((((((........(((.....))).........))))))))(((....)))..........)))..................... (-20.89 = -21.53 + 0.64)


| Location | 22,377,679 – 22,377,793 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -19.43 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22377679 114 - 23771897 GGGGGGGGGGUGUGUUGUUUUGCUCCUCCCCUUCGACAUGUGUCGCAUGUGGCUUUUCUCCGGCCUCACGCC-----ACCCCACAUGCAGCAUCAAGAUUAAUUCAUUCGCUUUUUGCU ((((((((((.(((......))).))))))))))(((....)))((((((((.........(((.....)))-----...))))))))((((..(((.............)))..)))) ( -41.42) >DroSec_CAF1 28756 100 - 1 -------------------UUGCUCCUCCCCUUCGACAUGUGUCGCAUGUGGCUUUUCUCCGGCCUCACGCCACUCCACCCCACAUGCAGCUUUAAGAUUAAUUCAUUCGCUUUUUGCU -------------------..(((..........(((....)))((((((((.........(((.....)))........)))))))))))..................((.....)). ( -22.83) >DroSim_CAF1 44299 100 - 1 -------------------UUGCUCCUCCCCUUCGACAUGUGUCGCAUGUGGCUUUUCUCCGGCCUCACGCCACUCCACCCCACAUGCAGCUUUAAGAUUAAUUCAUUCGCUUUUUGCU -------------------..(((..........(((....)))((((((((.........(((.....)))........)))))))))))..................((.....)). ( -22.83) >DroEre_CAF1 45168 93 - 1 ---------------------GCUCCUCCCCUGCGACAUGUGUCGCAAGUGGCCUUUCUCCGGCCUCCCGCC-----AGCCCACAUGCAGCUUUAAGAUUAAUUCAUUUGCUUUUUGCU ---------------------((........((((((....)))))).((((.((......(((.....)))-----)).))))..))(((...((((.(((.....))).)))).))) ( -20.90) >DroYak_CAF1 38599 95 - 1 -------------------UUGCUCCUCCCCUGCGACAUCUGUCGCUAGCGGCUUUCCUCCGGCCUCACGCU-----AACCCACAUGCAGCUUUAAGAUUAAUUCAUUCGCUUUUUGCU -------------------((((.........(((((....)))))((((((((.......))))....)))-----)........))))...................((.....)). ( -18.40) >consensus ___________________UUGCUCCUCCCCUUCGACAUGUGUCGCAUGUGGCUUUUCUCCGGCCUCACGCC_____ACCCCACAUGCAGCUUUAAGAUUAAUUCAUUCGCUUUUUGCU .....................(((..........(((....)))((((((((.........(((.....)))........)))))))))))..................((.....)). (-19.43 = -20.07 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:49 2006