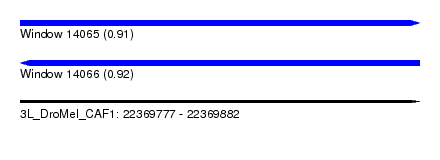
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,369,777 – 22,369,882 |
| Length | 105 |
| Max. P | 0.915385 |

| Location | 22,369,777 – 22,369,882 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.84 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
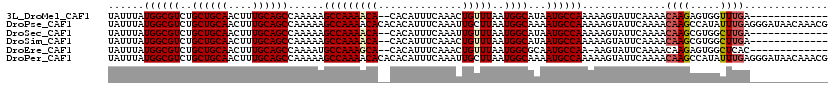
>3L_DroMel_CAF1 22369777 105 + 23771897 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACA--CACAUUUCAAACUGUUUAAUGGCAUAAUGCCAAAAAGUAUUCAAAACAAGAGUGGUUUGA------------- ......((((....((((((....))))))......)))).....--......((((((..(((..((((.....))))....((.......))..)))..))))))------------- ( -28.20) >DroPse_CAF1 71826 120 + 1 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACACACACAUUUCAAAUUGCUUAAUGGCAAAAUGCCAAAAAGUAUUCAAAACAAGCCAUAUUUGAGGGAUAACAAACG ......((((....((((((....))))))......))))...........((((((((.((((..((((.....))))....((.......))))))...))))))))........... ( -27.80) >DroSec_CAF1 20907 105 + 1 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACA--CACAUUUCAAAUUGUUUAAUGGCAUAAUGCCAAAAAGUAUUCAAAACAAGCGUGGCUUGA------------- ..(((.((((((..((((((....))))))......(((((((((--............)))))..))))...)))))).)))..........(((((...))))).------------- ( -27.10) >DroSim_CAF1 35294 105 + 1 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACA--CACAUUUCAAACUGUUUAAUGGCAUAAUGCCAAAAAGUAUUCAAAACAAGCGUGGCUUGA------------- ..(((.((((((..((((((....))))))......(((((((((--............)))))..))))...)))))).)))..........(((((...))))).------------- ( -27.10) >DroEre_CAF1 37920 104 + 1 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAUGCCAAAGCA--CACAUUUCAAACUGUUUAAUGGCGCAAUGCCAA-AAGUAUUCAAAACAAGAGUGGCUCAC------------- ......((((((..((((((....))))))....)))))).(((.--(((................((((.....)))).-..((.......))....))))))...------------- ( -26.40) >DroPer_CAF1 74498 120 + 1 UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACACACACAUUUCAAAUUGCUUAAUGGCAAAAUGCCAAAAAGUAUUCAAAACAAGCCAUAUUUGAGGGAUAACAAACG ......((((....((((((....))))))......))))...........((((((((.((((..((((.....))))....((.......))))))...))))))))........... ( -27.80) >consensus UAUUUAUGGCGUCUGCUGCAACUUUGCAGCCAAAAAGCCAAAACA__CACAUUUCAAACUGUUUAAUGGCAUAAUGCCAAAAAGUAUUCAAAACAAGCGUGGCUUGA_____________ ......((((((..((((((....))))))......(((((((((..............)))))..))))...))))))..............((((.....)))).............. (-21.51 = -21.84 + 0.33)
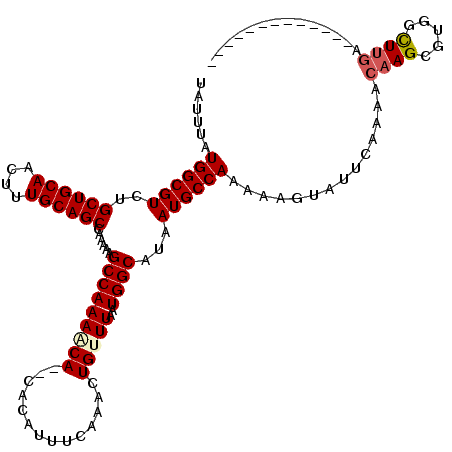

| Location | 22,369,777 – 22,369,882 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -25.47 |
| Energy contribution | -24.47 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
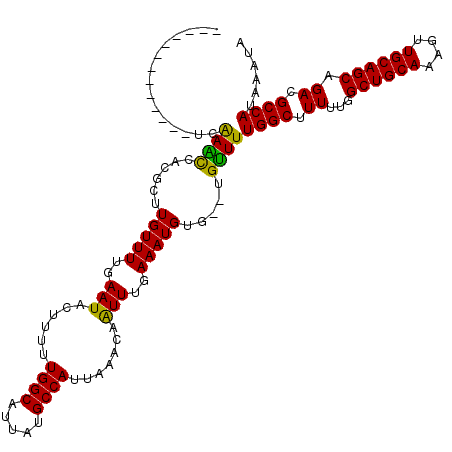
>3L_DroMel_CAF1 22369777 105 - 23771897 -------------UCAAACCACUCUUGUUUUGAAUACUUUUUGGCAUUAUGCCAUUAAACAGUUUGAAAUGUG--UGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA -------------..(((((((....((((..(((..((..((((.....))))..))...)))..)))))))--.))))((((.((...((((((....)))))).)).))))...... ( -28.20) >DroPse_CAF1 71826 120 - 1 CGUUUGUUAUCCCUCAAAUAUGGCUUGUUUUGAAUACUUUUUGGCAUUUUGCCAUUAAGCAAUUUGAAAUGUGUGUGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA .(((((........)))))(((((..((((..(((.(((..((((.....))))..)))..)))..))))..((.((((..((((....))))(((....))))))).)))))))..... ( -31.60) >DroSec_CAF1 20907 105 - 1 -------------UCAAGCCACGCUUGUUUUGAAUACUUUUUGGCAUUAUGCCAUUAAACAAUUUGAAAUGUG--UGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA -------------......(((((..((((..(((..((..((((.....))))..))...)))..)))))))--))...((((.((...((((((....)))))).)).))))...... ( -30.10) >DroSim_CAF1 35294 105 - 1 -------------UCAAGCCACGCUUGUUUUGAAUACUUUUUGGCAUUAUGCCAUUAAACAGUUUGAAAUGUG--UGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA -------------......(((((..((((..(((..((..((((.....))))..))...)))..)))))))--))...((((.((...((((((....)))))).)).))))...... ( -30.40) >DroEre_CAF1 37920 104 - 1 -------------GUGAGCCACUCUUGUUUUGAAUACUU-UUGGCAUUGCGCCAUUAAACAGUUUGAAAUGUG--UGCUUUGGCAUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA -------------..(((((((....((((..(((....-.((((.....)))).......)))..)))))))--.))))((((.((...((((((....)))))).)).))))...... ( -29.80) >DroPer_CAF1 74498 120 - 1 CGUUUGUUAUCCCUCAAAUAUGGCUUGUUUUGAAUACUUUUUGGCAUUUUGCCAUUAAGCAAUUUGAAAUGUGUGUGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA .(((((........)))))(((((..((((..(((.(((..((((.....))))..)))..)))..))))..((.((((..((((....))))(((....))))))).)))))))..... ( -31.60) >consensus _____________UCAAACCACGCUUGUUUUGAAUACUUUUUGGCAUUAUGCCAUUAAACAAUUUGAAAUGUG__UGUUUUGGCUUUUUGGCUGCAAAGUUGCAGCAGACGCCAUAAAUA ...............((((......(((((..(((......((((.....)))).......)))..))))).....))))((((.((...((((((....)))))).)).))))...... (-25.47 = -24.47 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:38 2006