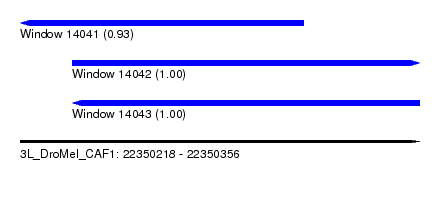
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,350,218 – 22,350,356 |
| Length | 138 |
| Max. P | 0.999721 |

| Location | 22,350,218 – 22,350,316 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22350218 98 - 23771897 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGAUGCCCUAC--CCCACU-------------------- (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))....((((((....))).))).....--......-------------------- ( -20.30) >DroPse_CAF1 49851 115 - 1 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGAGACGCCC-----CCGACUGGCGGCGGCAGCGGCAGUGG (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))....((((((....))).)))..-----((.((((.(.((....)).))))))) ( -32.40) >DroSec_CAF1 1176 100 - 1 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGAUGCCCCACAACCCACU-------------------- (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))..............(((.((.....)).)))...-------------------- ( -21.50) >DroSim_CAF1 14593 100 - 1 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUAAAUUUGCAUUUAAUUUGCGCUUACGAGGGAUGCCCCACAACCCACU-------------------- ...........((..(((((((((((((((((.....)))))).)))))))))))....(((.......)))......))(((.((.....)).)))...-------------------- ( -20.10) >DroEre_CAF1 18020 98 - 1 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGACGCCGCCC--CCUACU-------------------- (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))....((((((....))).))).....--......-------------------- ( -22.20) >DroPer_CAF1 52410 115 - 1 UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGAGACGCCC-----CCGACUGGCGGAGGCAGCGGCAGAGG (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))).((((((((..(....)..(((.-----(((.....))).)))))).))))).. ( -33.90) >consensus UAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGAGGGACGCCCCAC__CCCACU____________________ (((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).)))))....((((((....))).)))................................. (-20.91 = -20.47 + -0.44)


| Location | 22,350,236 – 22,350,356 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22350236 120 + 23771897 UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCACUUUAACGCUGAAGAGCUCGGCUUCACCA .(....).(((.......)))..((((((((((((..((((((.....)))))).....))))))))))))(((((..(((((((....)))))))..(((((.....)))))))))).. ( -31.60) >DroPse_CAF1 49886 120 + 1 UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCCAGUCCCAGU ..(.(((((((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).....(((..((((............))))..))))))))......... ( -26.90) >DroSec_CAF1 1196 120 + 1 UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCGGCUCCACCA ..(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)........(((((.....)))))....... ( -29.90) >DroSim_CAF1 14613 120 + 1 UCGUAAGCGCAAAUUAAAUGCAAAUUUAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCGGCUCCACCA ..(.((((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))).)........(((((.....)))))....... ( -26.60) >DroEre_CAF1 18038 120 + 1 UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCGGCUCCACCA ..(..(((((...(((.((.((.((((((((((((..((((((.....)))))).....)))))))))))).)).)).))).)))))..)........(((((.....)))))....... ( -29.90) >DroPer_CAF1 52445 120 + 1 UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCCAGUCCNNNN ..(.(((((((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).....(((..((((............))))..))))))))......... ( -26.90) >consensus UCGUGAGCGCAAAUUAAAUGCAAAUUCAAUUAAUUUGAAGCAAACAAUUUGCUUAAAUUAAUUAAUUGAAUGUGAAUCUAAAGUGCUUCCAAUUUAACGCUGAAGAGCUCGGCUCCACCA ....(((((((.......)))..((((((((((((..((((((.....)))))).....)))))))))))).....(((..((((............))))..))))))).......... (-25.64 = -25.67 + 0.03)


| Location | 22,350,236 – 22,350,356 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -28.41 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22350236 120 - 23771897 UGGUGAAGCCGAGCUCUUCAGCGUUAAAGUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA .((.....))((((..(((.((......)).)))(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..))))))).... ( -33.40) >DroPse_CAF1 49886 120 - 1 ACUGGGACUGGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA ..........((((..(((((.......))))).(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..))))))).... ( -29.70) >DroSec_CAF1 1196 120 - 1 UGGUGGAGCCGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA ..(((((((...))))...(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))))).. ( -32.30) >DroSim_CAF1 14613 120 - 1 UGGUGGAGCCGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUAAAUUUGCAUUUAAUUUGCGCUUACGA ..(.(((((...))))).)(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))..... ( -30.90) >DroEre_CAF1 18038 120 - 1 UGGUGGAGCCGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA ..(((((((...))))...(((((.((((((((.(((.....((.......))..(((((((((((((((((.....)))))).)))))))))))...))).)))))))))))))))).. ( -32.30) >DroPer_CAF1 52445 120 - 1 NNNNGGACUGGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA ..........((((..(((((.......))))).(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..))))))).... ( -29.70) >consensus UGGUGGAGCCGAGCUCUUCAGCGUUAAAUUGGAAGCACUUUAGAUUCACAUUCAAUUAAUUAAUUUAAGCAAAUUGUUUGCUUCAAAUUAAUUGAAUUUGCAUUUAAUUUGCGCUCACGA ..........((((..(((((.......))))).(((..((((((.((.((((((((((((.....((((((.....))))))..)))))))))))).)).))))))..))))))).... (-28.41 = -28.60 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:15 2006