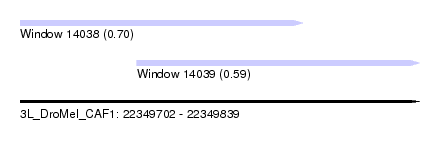
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,349,702 – 22,349,839 |
| Length | 137 |
| Max. P | 0.696608 |

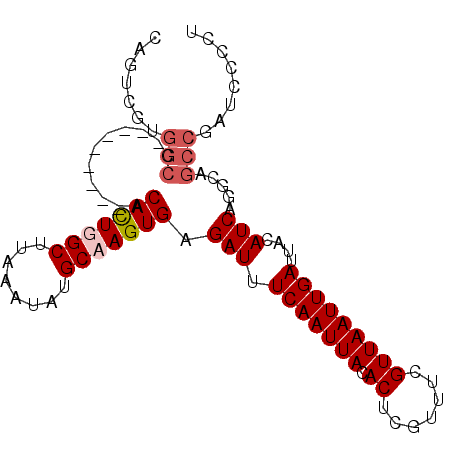
| Location | 22,349,702 – 22,349,799 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -11.64 |
| Energy contribution | -14.38 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22349702 97 + 23771897 GCCACAAAAUGUGCAGAACUUAAGGAUUAUUCAUUCUGCACAGUCGUGGCCAGGCCGCACACAUUUGCUUAAAUAUGCAAGUGAGAUUUCAAUUACA (((((....(((((((((......((....)).)))))))))...)))))...........(((((((........))))))).............. ( -28.90) >DroSec_CAF1 657 87 + 1 GCCACAGAAUUUGCAGAACU-AAGCAUUAUUCAUUCGACACAGUCGUGGCC---------ACAUUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACA (((((.((((.(((......-..)))..))))....(((...)))))))).---------.((((.((........)).)))).............. ( -19.50) >DroSim_CAF1 14098 87 + 1 GCCACAGAAUUUGCAGAACU-AAGCAUUAUUCAUUCGACACAGUCGUGGCC---------ACACUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACA (((((.((((.(((......-..)))..))))....(((...)))))))).---------.((((.((........)).)))).............. ( -21.80) >DroEre_CAF1 17576 77 + 1 GCCACAGAAGUUGCAGAACC-CAGCAUUAUUCAUGCUGCAC-------------------ACACUUGCUUAAAUAUGCAAGUGAGAUUUCAAUUACA ......((((((........-((((((.....))))))...-------------------.(((((((........))))))).))))))....... ( -21.20) >consensus GCCACAGAAUUUGCAGAACU_AAGCAUUAUUCAUUCGACACAGUCGUGGCC_________ACACUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACA (((((......(((((((...............))))))).....)))))...........(((((((........))))))).............. (-11.64 = -14.38 + 2.75)

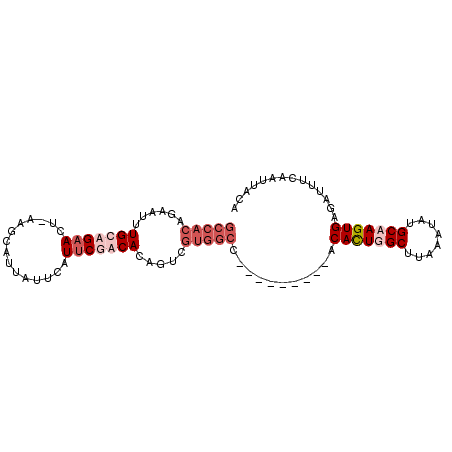

| Location | 22,349,742 – 22,349,839 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -14.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22349742 97 + 23771897 CAGUCGUGGCCAGGCCGCACACAUUUGCUUAAAUAUGCAAGUGAGAUUUCAAUUACACUCGUUUCGUUAAUUGAUUACAUCAGGCUGCCGAUCCCCU ((((((((((...)))))...(((((((........))))))).(((.(((((((.((.......)))))))))....))).))))).......... ( -25.80) >DroSec_CAF1 696 88 + 1 CAGUCGUGGCC---------ACAUUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACACUCGUUUCGUUAAUUGAUUACAUCAGGCAGCCGAUCCCCU ..((((..(((---------.((((.((........)).)))).(((.(((((((.((.......)))))))))....))).)))...))))..... ( -18.20) >DroSim_CAF1 14137 88 + 1 CAGUCGUGGCC---------ACACUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACACUCGUUUCGUUAAUUGAUUACAUCAGGCAGCCGAUCCCCU ..((((..(((---------.((((.((........)).)))).(((.(((((((.((.......)))))))))....))).)))...))))..... ( -20.50) >DroEre_CAF1 17615 78 + 1 C-------------------ACACUUGCUUAAAUAUGCAAGUGAGAUUUCAAUUACACUCGUUUCGUUAAUUGAUUACAUCAGGCAAGCGAUCCCCU .-------------------.(((((((........))))))).................(..(((((..(((((...)))))...)))))..)... ( -17.40) >consensus CAGUCGUGGCC_________ACACUGGCUUAAAUAUGCAAGUGAGAUUUCAAUUACACUCGUUUCGUUAAUUGAUUACAUCAGGCAGCCGAUCCCCU .......(((...........(((((((........))))))).(((.(((((((.((.......)))))))))....))).....)))........ (-14.20 = -15.20 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:12 2006