
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,335,674 – 22,335,847 |
| Length | 173 |
| Max. P | 0.973023 |

| Location | 22,335,674 – 22,335,790 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -16.18 |
| Energy contribution | -15.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22335674 116 + 23771897 CCCCAUACCAUCUUGCAAG---CGCGACUCUGUCUCGUAAAAGGCCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGU ..(((((.......(((((---((.((((((((((.((.....)).))..)))))....)))....))))).)).......(((((((-......)))))))........)))))..... ( -25.70) >DroPse_CAF1 32776 106 + 1 CCCUAGACUG---UGGAGGGUGGGGGGGAGUGGCU---------CUAGCCACGAAAAA--AAAUUACGCUUAGUUUUUACGGCUUCUGGAGAACCCAGAAGCAUAAUUUUUAUGGUCCAU (((((..((.---...))..))))).((((((((.---------...)))))....((--((((((......((....)).((((((((.....)))))))).))))))))....))).. ( -37.40) >DroSim_CAF1 3212 116 + 1 CCCCAUACCAUAUCGCAAG---CGCGACUCUGUCUCGUAAAAGGCCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGU .......(((((..(((((---((.((((((((((.((.....)).))..)))))....)))....))))).)).......(((((((-......)))))))........)))))..... ( -26.10) >DroEre_CAF1 3228 116 + 1 CCCCAUACCAUCUCGCAAG---CGCGACUCUGCCUCAUAAAUGACCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGU ..(((((.......(((((---((((....)))......((((((.((........)).))))))..)))).)).......(((((((-......)))))))........)))))..... ( -23.60) >DroPer_CAF1 33170 105 + 1 CCCUAGACUG---UGGAAG---GGGGGGAGUGGCU---------CUAGCCACGAAAUUUGUCAUUACGCUUAGUUUUUACGGCUUCUGGAGAACCCAGAAGCAUAAUUUUUAUGGUCCAU .....(((((---((((((---(((.(..(((((.---------...)))))((......))....).)))..........((((((((.....))))))))....)))))))))))... ( -33.40) >consensus CCCCAUACCAU_UUGCAAG___CGCGACUCUGGCUC_UAAA_G_CCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG_AAAACCCAGAAGCAUAAUUUUUAUGGCCCGU ...............................................((((.((((..(((.(..((.....))..).)))(((((((.......))))))).....)))).)))).... (-16.18 = -15.94 + -0.24)

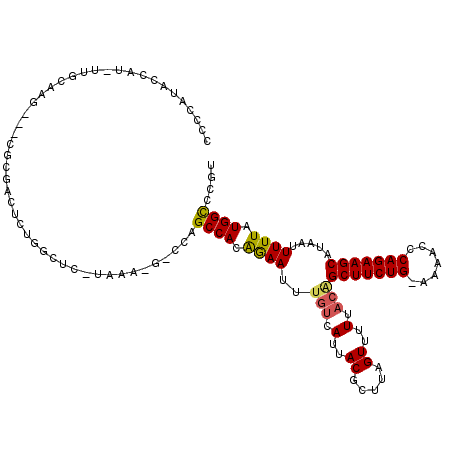
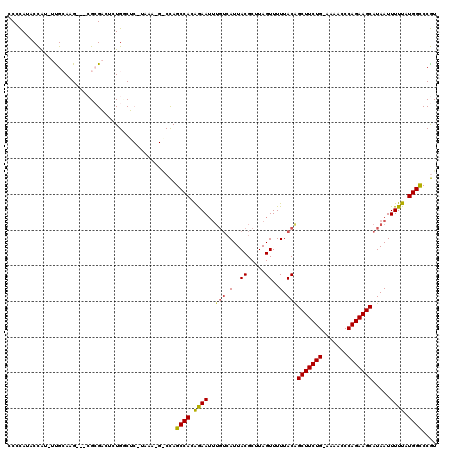
| Location | 22,335,711 – 22,335,813 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22335711 102 + 23771897 AAGGCCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGG--------------ACCCU---CGGGCAACCCUU ..(((((.....(((((((((.(..((.....))..).)))(((((((-......)))))))..))))))..))))).(((((((.--------------...))---)))))....... ( -31.60) >DroPse_CAF1 32808 112 + 1 ----CUAGCCACGAAAAA--AAAUUACGCUUAGUUUUUACGGCUUCUGGAGAACCCAGAAGCAUAAUUUUUAUGGUCCAU--GGAAGCCUUUUUUCCCUUACCCUUCGCUGUUAACCCUU ----.((((..((((.((--((((((......((....)).((((((((.....)))))))).))))))))..(((....--(((((.....)))))...))).))))..))))...... ( -26.10) >DroSim_CAF1 3249 102 + 1 AAGGCCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGG--------------ACCCU---CGGGCAACCCUU ..(((((.....(((((((((.(..((.....))..).)))(((((((-......)))))))..))))))..))))).(((((((.--------------...))---)))))....... ( -31.60) >DroEre_CAF1 3265 102 + 1 AUGACCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG-AAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGG--------------ACCCU---CGGGCAACCCUU ..(((..((((.((((..(((.(..((.....))..).)))(((((((-......))))))).....)))).))))..))).(((.--------------.((..---..))...))).. ( -29.30) >DroPer_CAF1 33199 114 + 1 ----CUAGCCACGAAAUUUGUCAUUACGCUUAGUUUUUACGGCUUCUGGAGAACCCAGAAGCAUAAUUUUUAUGGUCCAU--GGAAGCCUUUUUCCCCUUACCCUUCGCUGUUAACCCUU ----.((((..((((...(((.(..((.....))..).)))((((((((.....))))))))...........(((....--(((((....)))))....))).))))..))))...... ( -24.10) >consensus A_G_CCAGCCACAGAAUUUGUCAUUACGCUUAGUUUUUACAGCUUCUG_AAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGG______________ACCCU___CGGGCAACCCUU .......((((.((((..(((.(..((.....))..).)))(((((((.......))))))).....)))).))))......(((................))).....((....))... (-16.83 = -17.39 + 0.56)

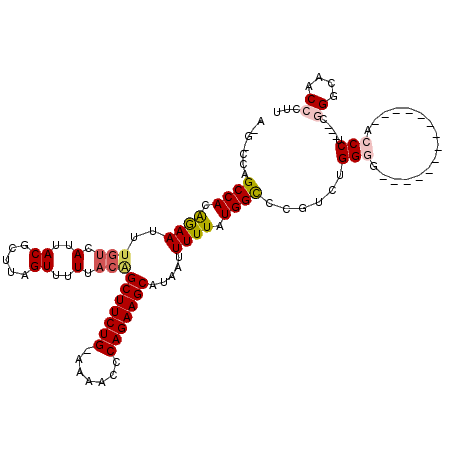

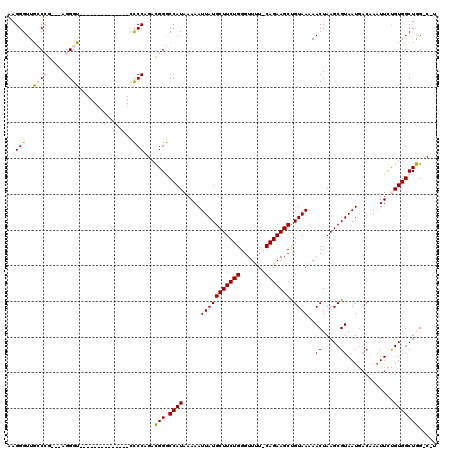
| Location | 22,335,711 – 22,335,813 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -17.97 |
| Energy contribution | -17.81 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22335711 102 - 23771897 AAGGGUUGCCCG---AGGGU--------------CCCCAGACGGGCCAUAAAAAUUAUGCUUCUGGGUUUU-CAGAAGCUGUAAAAACUAAGCGUAAUGACAAAUUCUGUGGCUGGCCUU ..(((..(((..---..)))--------------.)))((.(.(((((((........((((((((....)-)))))))(((....((.....))....))).....))))))).).)). ( -30.50) >DroPse_CAF1 32808 112 - 1 AAGGGUUAACAGCGAAGGGUAAGGGAAAAAAGGCUUCC--AUGGACCAUAAAAAUUAUGCUUCUGGGUUCUCCAGAAGCCGUAAAAACUAAGCGUAAUUU--UUUUUCGUGGCUAG---- ...((((....(((((((((...((((.......))))--....)))..(((((((((((((((((.....)))))))).((....)).....)))))))--))))))))))))..---- ( -28.30) >DroSim_CAF1 3249 102 - 1 AAGGGUUGCCCG---AGGGU--------------CCCCAGACGGGCCAUAAAAAUUAUGCUUCUGGGUUUU-CAGAAGCUGUAAAAACUAAGCGUAAUGACAAAUUCUGUGGCUGGCCUU ..(((..(((..---..)))--------------.)))((.(.(((((((........((((((((....)-)))))))(((....((.....))....))).....))))))).).)). ( -30.50) >DroEre_CAF1 3265 102 - 1 AAGGGUUGCCCG---AGGGU--------------CCCCAGACGGGCCAUAAAAAUUAUGCUUCUGGGUUUU-CAGAAGCUGUAAAAACUAAGCGUAAUGACAAAUUCUGUGGCUGGUCAU ..(((..(((..---..)))--------------.))).(((.(((((((........((((((((....)-)))))))(((....((.....))....))).....))))))).))).. ( -35.40) >DroPer_CAF1 33199 114 - 1 AAGGGUUAACAGCGAAGGGUAAGGGGAAAAAGGCUUCC--AUGGACCAUAAAAAUUAUGCUUCUGGGUUCUCCAGAAGCCGUAAAAACUAAGCGUAAUGACAAAUUUCGUGGCUAG---- ...((((....(((...(((....((((......))))--....)))...........((((((((.....)))))))))))...)))).(((...((((......)))).)))..---- ( -27.90) >consensus AAGGGUUGCCCG___AGGGU______________CCCCAGACGGGCCAUAAAAAUUAUGCUUCUGGGUUUU_CAGAAGCUGUAAAAACUAAGCGUAAUGACAAAUUCUGUGGCUGG_C_U ..(((..((((.....))))...............)))...(((.((((.....(((((((((((.......))))))).))))..((.....)).............)))))))..... (-17.97 = -17.81 + -0.16)


| Location | 22,335,751 – 22,335,847 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22335751 96 + 23771897 AGCUUCUGAAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGGACCCUCGGGCAACCCUUUGUCCCACCCACCCCCUGCUUGCAGCACUGGU----UG .(((((((......)))))))...........((((.((((((((((....((....))....)))))...((.....))....))).)).)))----). ( -29.80) >DroSim_CAF1 3289 100 + 1 AGCUUCUGAAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGGACCCUCGGGCAACCCUUUGUCCCACCCACCCCCUGCUUGCAGCACUGGUUGGUUG .(((((((......)))))))............(((((...((....)).)))))............((.(((.((..(((.....)))..)).))).)) ( -33.10) >DroEre_CAF1 3305 95 + 1 AGCUUCUGAAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGGACCCUCGGGCAACCCUUUGUGCCACCCACC-CCUGCUUGCAGCACUGCU----UG .(((((((......)))))))............(((((...((....)).)))))........((((((..((..-..))..))..))))....----.. ( -27.70) >consensus AGCUUCUGAAAACCCAGAAGCAUAAUUUUUAUGGCCCGUCUGGGGACCCUCGGGCAACCCUUUGUCCCACCCACCCCCUGCUUGCAGCACUGGU____UG .(((((((......)))))))............(((((...((....)).)))))...............(((....(((....)))...)))....... (-26.40 = -26.73 + 0.33)

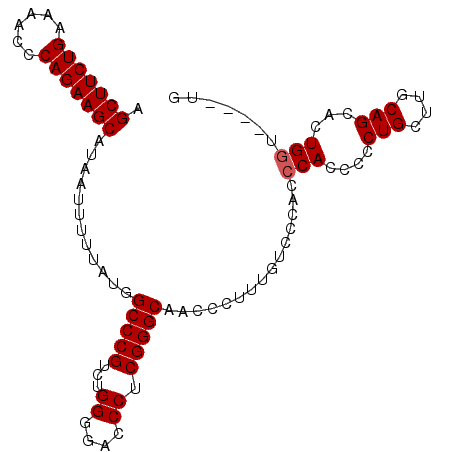
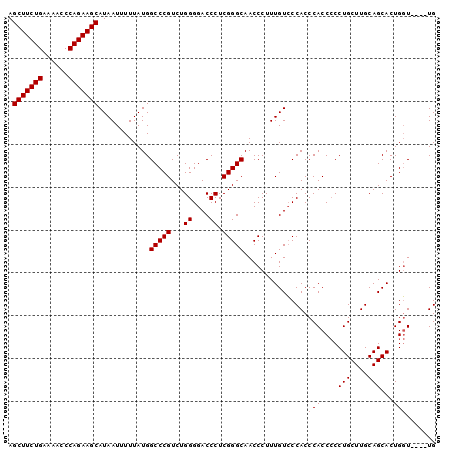
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:19:05 2006