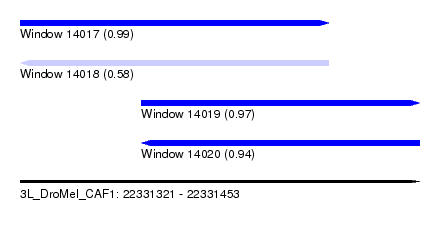
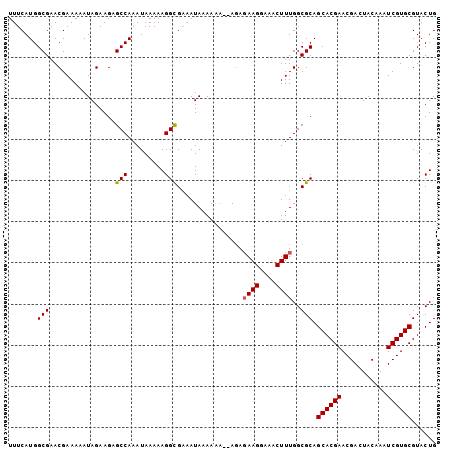
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,331,321 – 22,331,453 |
| Length | 132 |
| Max. P | 0.993639 |

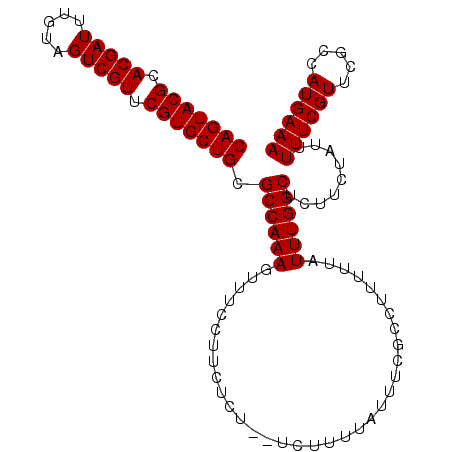
| Location | 22,331,321 – 22,331,423 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.98 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22331321 102 + 23771897 CAGUACGCACGAUUUGUAGUCGUUCGUGCUGCGCCAAAGUUUCCUUCUCU--UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAA (((((((.(((((.....))))).))))))).((((((............--....................)))))).........((((((.....)))))) ( -21.65) >DroSec_CAF1 10668 101 + 1 CAGUACGCACGAUUUGUAGUCGUUCGUGCUGCGCCAAAGUUU-CUUCUCU--UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAA (((((((.(((((.....))))).))))))).((((((....-.......--....................)))))).........((((((.....)))))) ( -21.68) >DroEre_CAF1 8599 101 + 1 CAGUACGCACGAUUUGUAGUCGUUCGUGCUGCGCCAAAGUUUCCUUCUCU---UUUUUAUUCCACCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAA (((((((.(((((.....))))).))))))).((((((............---...................)))))).........((((((.....)))))) ( -21.68) >DroYak_CAF1 8014 104 + 1 CAGUACGCACGAUUUGUAGUCGUUCGUGCUGCGCCAAAGUUUCCUUAUAUUUUUUUUUAUAUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAA (((((((.(((((.....))))).))))))).((((((.......((((........))))...........)))))).........((((((.....)))))) ( -22.77) >consensus CAGUACGCACGAUUUGUAGUCGUUCGUGCUGCGCCAAAGUUUCCUUCUCU__UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAA (((((((.(((((.....))))).))))))).((((((..................................)))))).........((((((.....)))))) (-21.58 = -21.58 + 0.00)

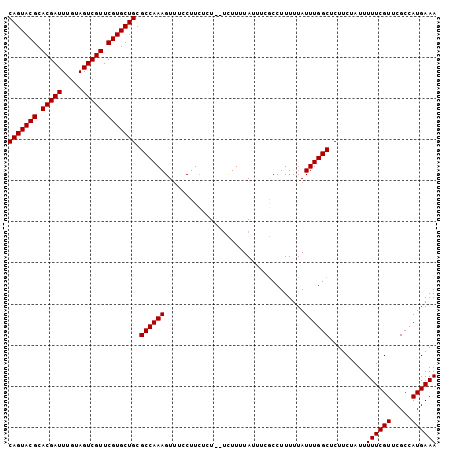
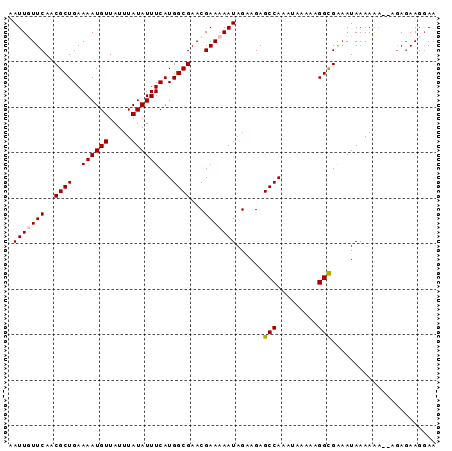
| Location | 22,331,321 – 22,331,423 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 94.98 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22331321 102 - 23771897 UUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAGA--AGAGAAGGAAACUUUGGCGCAGCACGAACGACUACAAAUCGUGCGUACUG .......(((................(((.........)))...........--...((((....))))..))).((((((...........))))))...... ( -20.60) >DroSec_CAF1 10668 101 - 1 UUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAGA--AGAGAAG-AAACUUUGGCGCAGCACGAACGACUACAAAUCGUGCGUACUG .....((((.................)))).........(((.....((((.--.......-...))))..))).((((((...........))))))...... ( -16.33) >DroEre_CAF1 8599 101 - 1 UUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGUGGAAUAAAAA---AGAGAAGGAAACUUUGGCGCAGCACGAACGACUACAAAUCGUGCGUACUG .....((((.................))))........((((.........---.(.((((....))))..)...((((((...........)))))).)))). ( -19.13) >DroYak_CAF1 8014 104 - 1 UUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAUAUAAAAAAAAAUAUAAGGAAACUUUGGCGCAGCACGAACGACUACAAAUCGUGCGUACUG .......(((................(((.........))).................(((....)))...))).((((((...........))))))...... ( -18.60) >consensus UUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAAA__AGAGAAGGAAACUUUGGCGCAGCACGAACGACUACAAAUCGUGCGUACUG .......(((................(((.........)))................((((....))))..))).((((((...........))))))...... (-18.04 = -18.10 + 0.06)

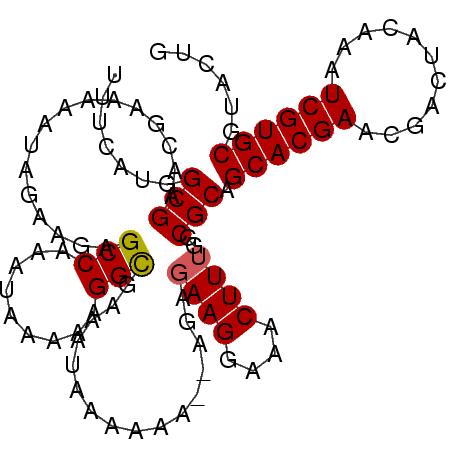
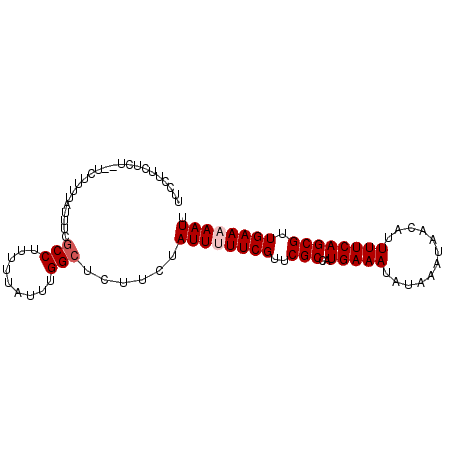
| Location | 22,331,361 – 22,331,453 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -11.15 |
| Consensus MFE | -10.30 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22331361 92 + 23771897 UUCCUUCUCU--UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAAUAUAAAUAACAUUUUCAGCGUUGAAAAAUU ..........--...........(((.........)))......((((((((..(((..(((((............)))))))).)))))))). ( -13.30) >DroSec_CAF1 10708 91 + 1 UU-CUUCUCU--UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAAUAUAAAUAACAUUUUCAGCGUUGAAAAAUU ..-.......--...........(((.........)))......((((((((..(((..(((((............)))))))).)))))))). ( -13.30) >DroEre_CAF1 8639 91 + 1 UUCCUUCUCU---UUUUUAUUCCACCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAAUAUAAAUAACAUUUUCAGCGUUGAACAAUU ..........---......................................(((((.(.(((((............)))))..).))))).... ( -7.10) >DroYak_CAF1 8054 94 + 1 UUCCUUAUAUUUUUUUUUAUAUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAAUAUAAAUAACAUUUUCAGCGUUGAACAAUU .......................(((.........))).............(((((.(.(((((............)))))..).))))).... ( -10.90) >consensus UUCCUUCUCU__UCUUUUAUUUCGCCUUUUUAUUUGGCUCUUCUAUUUUUCGUUCGCCAUGAAAUAUAAAUAACAUUUUCAGCGUUGAAAAAUU .......................(((.........)))......((((((((..(((..(((((............)))))))).)))))))). (-10.30 = -11.05 + 0.75)


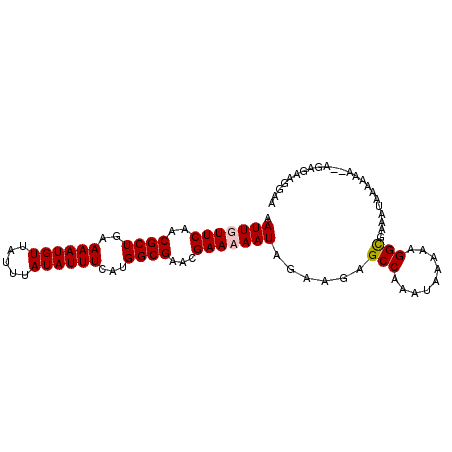
| Location | 22,331,361 – 22,331,453 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22331361 92 - 23771897 AAUUUUUCAACGCUGAAAAUGUUAUUUAUAUUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAGA--AGAGAAGGAA .(((((((..((((..((((((.....))))))...))))...)))))))......(((.........)))...........--.......... ( -13.70) >DroSec_CAF1 10708 91 - 1 AAUUUUUCAACGCUGAAAAUGUUAUUUAUAUUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAGA--AGAGAAG-AA .(((((((..((((..((((((.....))))))...))))...)))))))......(((.........)))...........--.......-.. ( -13.70) >DroEre_CAF1 8639 91 - 1 AAUUGUUCAACGCUGAAAAUGUUAUUUAUAUUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGUGGAAUAAAAA---AGAGAAGGAA ..((((((..((((..((((((.....))))))...))))................(((.........))).))))))...---.......... ( -12.00) >DroYak_CAF1 8054 94 - 1 AAUUGUUCAACGCUGAAAAUGUUAUUUAUAUUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAUAUAAAAAAAAAUAUAAGGAA ..((((((..((.(((((.(((.....))))))))))..))))))...........(((.........)))....................... ( -12.60) >consensus AAUUGUUCAACGCUGAAAAUGUUAUUUAUAUUUCAUGGCGAACGAAAAAUAGAAGAGCCAAAUAAAAAGGCGAAAUAAAAAA__AGAGAAGGAA .(((((((..((((..((((((.....))))))...))))...)))))))......(((.........)))....................... (-11.91 = -12.22 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:54 2006