
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,330,785 – 22,330,889 |
| Length | 104 |
| Max. P | 0.995686 |

| Location | 22,330,785 – 22,330,889 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -21.58 |
| Energy contribution | -23.82 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330785 104 + 23771897 GUCCACCCACACAGCGCUUUCCAACCCC-AAGGGCAUGCUCCAAUCCCACCCAGGUUAACAGCGGGAAUUGGAACUGGAACGGGAACGGGACACGUGUCGGUGGC ..(((((.((((......(((((.(((.-..))).....(((((((((.(...........).))).))))))..)))))((....))......)))).))))). ( -33.40) >DroSec_CAF1 10138 105 + 1 GUCCACCCACACAGCGCUUUCCAACCCCGAAGGGCAUGCUCCAAUCCCAGCCAGGUUAACAGCGGGAACUGGAACUGGAACGAGAACGGGACACGUGUCGGUGGC ..(((((.((((.((.(((((.......))))))).((.(((....((((((((..............))))..))))..((....))))))).)))).))))). ( -32.44) >DroEre_CAF1 8063 86 + 1 GUCCACCCACACAGCACUUUCCAACCCC-AGGGGUAUGCUCCAAUCCCAGCCAGGUUAACAAC------------------UGGAACGGGACACGUGUCGGUGGC ..(((((.((((((((..((((......-.))))..))))....((((..((((........)------------------)))...))))...)))).))))). ( -33.60) >DroYak_CAF1 7534 86 + 1 GUCCACCCACACAGCACUUUCCAACCCC-AAGGGAAUGCUCCAAUCCAACCCAGGUUAACAAU------------------UGGAACGGGACACGUGUCGGUGGC ..(((((.((((((((.(((((......-..)))))))))....(((...((((........)------------------)))....)))...)))).))))). ( -27.60) >consensus GUCCACCCACACAGCACUUUCCAACCCC_AAGGGCAUGCUCCAAUCCCACCCAGGUUAACAAC__________________GGGAACGGGACACGUGUCGGUGGC ..(((((.((((............(((....)))..((..((..((((..((((....................))))...))))..))..)).)))).))))). (-21.58 = -23.82 + 2.25)

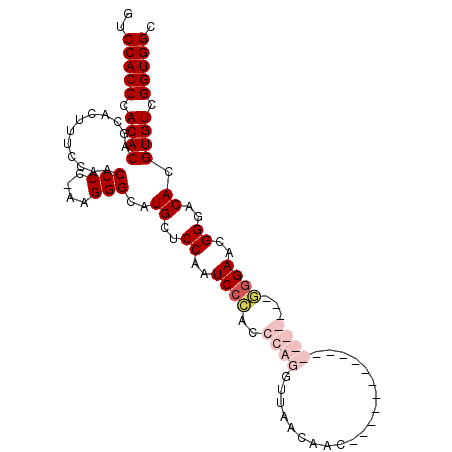
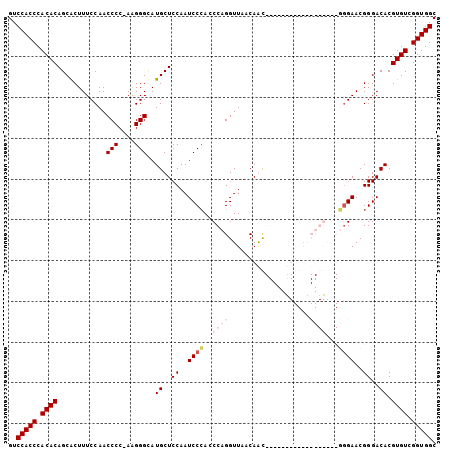
| Location | 22,330,785 – 22,330,889 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -30.95 |
| Energy contribution | -30.89 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330785 104 - 23771897 GCCACCGACACGUGUCCCGUUCCCGUUCCAGUUCCAAUUCCCGCUGUUAACCUGGGUGGGAUUGGAGCAUGCCCUU-GGGGUUGGAAAGCGCUGUGUGGGUGGAC .(((((.((((((((.(((..((((...((((((((((.((((((.........)))))))))))))).))....)-)))..)))...)))).)))).))))).. ( -49.60) >DroSec_CAF1 10138 105 - 1 GCCACCGACACGUGUCCCGUUCUCGUUCCAGUUCCAGUUCCCGCUGUUAACCUGGCUGGGAUUGGAGCAUGCCCUUCGGGGUUGGAAAGCGCUGUGUGGGUGGAC .(((((.((((((((..((....))((((((((((((((((.(((........))).)))))))))))..((((....))))))))).)))).)))).))))).. ( -53.50) >DroEre_CAF1 8063 86 - 1 GCCACCGACACGUGUCCCGUUCCA------------------GUUGUUAACCUGGCUGGGAUUGGAGCAUACCCCU-GGGGUUGGAAAGUGCUGUGUGGGUGGAC .(((((.((((...(((.((((((------------------(((........))))))))).)))((((((((..-.))))......)))).)))).))))).. ( -37.70) >DroYak_CAF1 7534 86 - 1 GCCACCGACACGUGUCCCGUUCCA------------------AUUGUUAACCUGGGUUGGAUUGGAGCAUUCCCUU-GGGGUUGGAAAGUGCUGUGUGGGUGGAC .(((((.((((((.(((.((((((------------------(((..((((....)))))))))))))..(((...-.)))..)))....)).)))).))))).. ( -35.30) >consensus GCCACCGACACGUGUCCCGUUCCA__________________GCUGUUAACCUGGCUGGGAUUGGAGCAUGCCCUU_GGGGUUGGAAAGCGCUGUGUGGGUGGAC .(((((.(((((((..(((.((((.................................)))).)))..)))((((....))))...........)))).))))).. (-30.95 = -30.89 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:51 2006