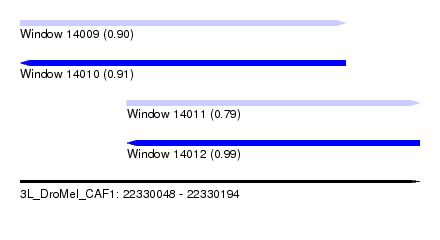
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,330,048 – 22,330,194 |
| Length | 146 |
| Max. P | 0.987883 |

| Location | 22,330,048 – 22,330,167 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330048 119 + 23771897 CAAGCCAAUCGGAAUUGCCCUUACUGCGGUCGGCU-UUGAGUUGGUUGGCAUGUAAUGCAGAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAA ......((((.(((..(((....((((((((((((-.......)))))))......))))).........((.(((.((((((.....))))))...)))...)))))..))).)))).. ( -34.10) >DroSec_CAF1 9413 119 + 1 GAAGCCAAUCUGAAUUGCCCUUGCUGCGGUCGGCUUUUGAGUUGGUUAGCAUGUAAUGCAGAUGUUU-CAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAA ((((((.(((((.(((((...(((((...((((((....)))))).))))).))))).)))))....-..((.(((.((((((.....))))))...)))...))))))))......... ( -36.20) >DroEre_CAF1 7349 104 + 1 UAAGCCAAUCUGAAUUGCCCUUACUGCGGUCG----------------GCAUGUAAUGCAUAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAA ......((((((((..(((........((..(----------------((((((.....))))))).)).((.(((.((((((.....))))))...)))...)))))..)))))))).. ( -31.20) >DroYak_CAF1 6840 104 + 1 UAAGCCAAUCUGAAUUGCCCUUACUGCGGUUG----------------GCAUGUAAUGCAUAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAA ......((((((((..(((........(((((----------------((((((.....)))))...))))))(((.((((((.....))))))...))).....)))..)))))))).. ( -34.00) >consensus UAAGCCAAUCUGAAUUGCCCUUACUGCGGUCG________________GCAUGUAAUGCAGAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAA ......((((((((..(((.(((((((.....................))).))))..............((.(((.((((((.....))))))...)))...)))))..)))))))).. (-26.66 = -26.72 + 0.06)

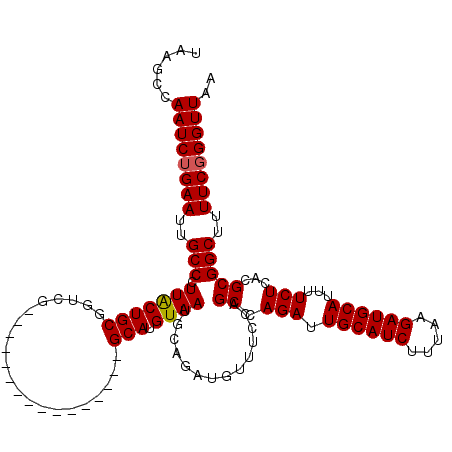

| Location | 22,330,048 – 22,330,167 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330048 119 - 23771897 UUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUCUGCAUUACAUGCCAACCAACUCAA-AGCCGACCGCAGUAAGGGCAAUUCCGAUUGGCUUG ..........((((((((.((((...((((((.....)))))).)))(((((....)....((((...))))............-))))..)))).....(((....)))....))))). ( -30.20) >DroSec_CAF1 9413 119 - 1 UUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUG-AAACAUCUGCAUUACAUGCUAACCAACUCAAAAGCCGACCGCAGCAAGGGCAAUUCAGAUUGGCUUC ..........(((((((((((((...((((((.....)))))).))).((.(-(....)).))....)))))..............(((............)))..........))))). ( -27.00) >DroEre_CAF1 7349 104 - 1 UUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC----------------CGACCGCAGUAAGGGCAAUUCAGAUUGGCUUA ..........(((((.(....).....(((((.....)))))(((((((.((....))..(((.((((.(((----------------.....)))))))..)))..)))))))))))). ( -31.30) >DroYak_CAF1 6840 104 - 1 UUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC----------------CAACCGCAGUAAGGGCAAUUCAGAUUGGCUUA ..........(((((.(....).....(((((.....)))))(((((((.((....))..(((.((((.(((----------------.....)))))))..)))..)))))))))))). ( -31.30) >consensus UUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC________________CGACCGCAGUAAGGGCAAUUCAGAUUGGCUUA ..........(((((.(....).....(((((.....)))))(((((((.((....))..(((.((((.(((.....................)))))))..)))..)))))))))))). (-25.25 = -25.75 + 0.50)

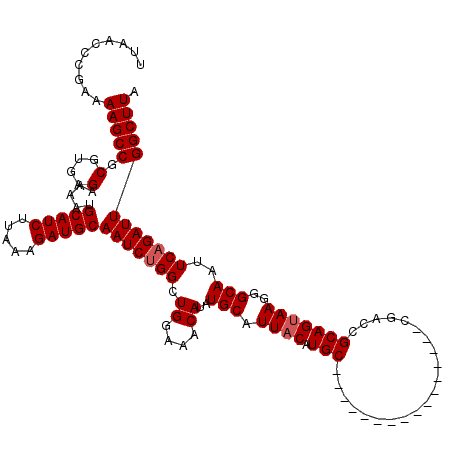
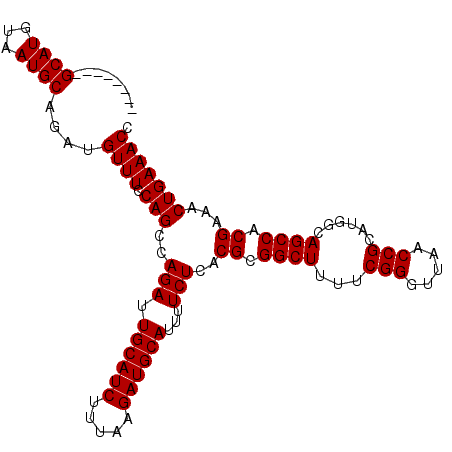
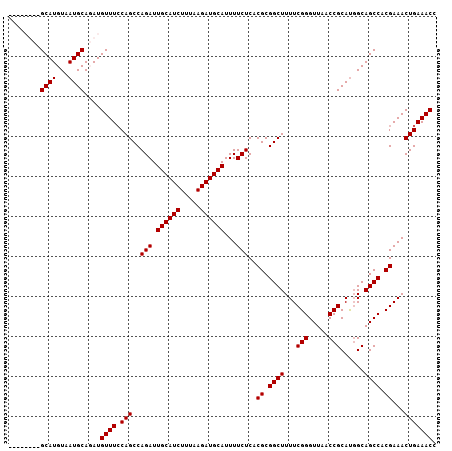
| Location | 22,330,087 – 22,330,194 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330087 107 + 23771897 GUUGGUUGGCAUGUAAUGCAGAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAACCGCAUGGCAGCCACGAAACUGAAACC ..((((((.(((((...............(((((((.((((((.....))))))...))).....))))....((.....))))))).))))))............. ( -31.90) >DroSec_CAF1 9453 106 + 1 GUUGGUUAGCAUGUAAUGCAGAUGUUU-CAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAACCGCAUGGCAGCCACGAAACUGAAACC ....(((.((((...)))).)))((((-(((..(((.((((((.....))))))...)))..((.((((...(((.....)))......)))).))...))))))). ( -30.30) >DroEre_CAF1 7381 99 + 1 --------GCAUGUAAUGCAUAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAACCGCACGGCAGCCACGAAACUGAAACC --------((((...))))....(((((.(((((((.((((((.....))))))...))).....))))(((((((((..((....)).)))).)))))..))))). ( -28.00) >DroYak_CAF1 6872 99 + 1 --------GCAUGUAAUGCAUAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAACCGCAUUGCAGCCACGAAACUGAAACC --------((.((((((((..........(((((((.((((((.....))))))...))).....))))....((.....))))))))))))............... ( -28.30) >consensus ________GCAUGUAAUGCAGAUGUUUCCAGCCAGAUUGCAUCUUUAAGAUGCAUUUUCUCACGCGGCUUUUCGGGUUAACCGCAUGGCAGCCACGAAACUGAAACC ........((((...))))....((((.(((..(((.((((((.....))))))...)))..((.((((...(((.....)))......)))).))...))))))). (-26.05 = -26.05 + 0.00)

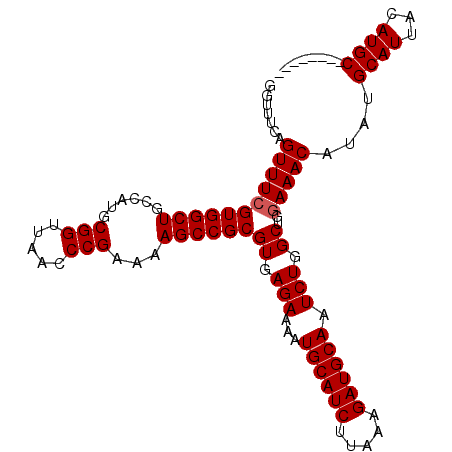
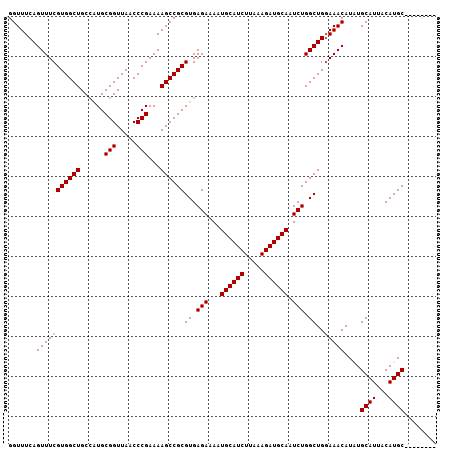
| Location | 22,330,087 – 22,330,194 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22330087 107 - 23771897 GGUUUCAGUUUCGUGGCUGCCAUGCGGUUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUCUGCAUUACAUGCCAACCAAC ((((...(((((((((((......(((.....)))...))))))((.(((...((((((.....)))))).))).))..)))))....((((...)))).))))... ( -35.40) >DroSec_CAF1 9453 106 - 1 GGUUUCAGUUUCGUGGCUGCCAUGCGGUUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUG-AAACAUCUGCAUUACAUGCUAACCAAC ((((...(((((((((((......(((.....)))...))))))((.(((...((((((.....)))))).))).)).)-))))....((((...)))).))))... ( -35.70) >DroEre_CAF1 7381 99 - 1 GGUUUCAGUUUCGUGGCUGCCGUGCGGUUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC-------- .......(((((((((((......(((.....)))...))))))((.(((...((((((.....)))))).))).))..)))))....((((...))))-------- ( -31.20) >DroYak_CAF1 6872 99 - 1 GGUUUCAGUUUCGUGGCUGCAAUGCGGUUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC-------- .......(((((((((((......(((.....)))...))))))((.(((...((((((.....)))))).))).))..)))))....((((...))))-------- ( -31.20) >consensus GGUUUCAGUUUCGUGGCUGCCAUGCGGUUAACCCGAAAAGCCGCGUGAGAAAAUGCAUCUUAAAGAUGCAAUCUGGCUGGAAACAUAUGCAUUACAUGC________ .......(((((((((((......(((.....)))...))))))((.(((...((((((.....)))))).))).))..)))))....((((...))))........ (-30.30 = -30.55 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:47 2006