
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,320,057 – 22,320,328 |
| Length | 271 |
| Max. P | 0.945849 |

| Location | 22,320,057 – 22,320,152 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -42.34 |
| Consensus MFE | -30.35 |
| Energy contribution | -33.23 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22320057 95 - 23771897 -------------------CAUCCCAGUGUAGGGCAGCGGGAAGAUGAGACCCAAAAGCGAGCCCGGCAGUCUUAUCUCGUCCCGUACAUCGGGCUUUAACUUUUGGCUGGCAC------ -------------------.......((((..(((.(((((((((((((((((............))..)))))))))..)))))).((..((.......))..))))).))))------ ( -28.60) >DroSec_CAF1 78731 120 - 1 UGCUUCGGCCAUCUCCAUUCAGCCCAGUGUAGGGCAGCGGGAAGAUGAGACCCAGAAGCGAGCCUGGCAGUCUUAUCUCGUCCCGUACAUCGGGCUUUAGCUUUUGGCUGGCACUCUGCA (((..((((((.........(((((.((((......(((((((((((((((((((........))))..)))))))))..)))))))))).)))))........)))))))))....... ( -47.33) >DroEre_CAF1 77099 120 - 1 UGGUUCGGCCAUCGCCAUUCAGCCCAGUGUAGGGCAGCGGGAAGAUGAGACCCAAAAGCGAGCCUGCCAGUCUUAUCUCGUCCCCUACAUCGGGCUUUAGCUUUUGCCUGGCACUCCGCA (((.....)))..((((...(((((.((((((((..(((...(((((((((......(((....)))..)))))))))))).)))))))).)))))...((....)).))))........ ( -44.10) >DroYak_CAF1 57120 120 - 1 UAGUUCGGCCAUCUCCAUUCAGCCCAGUGUAGGGCAGAGGGAAGAUGAGACCCAACAGCGAGCCUGGCAGUCUUAUCUUGUCCCCUGCAUCGGGCUUUAGCUUUUGGCUGGCACUCCGCA .((((((((((.........(((((.((((((((....((.((((((((((....(((.....)))...)))))))))).)))))))))).)))))........))))))).)))..... ( -49.33) >consensus UGGUUCGGCCAUCUCCAUUCAGCCCAGUGUAGGGCAGCGGGAAGAUGAGACCCAAAAGCGAGCCUGGCAGUCUUAUCUCGUCCCCUACAUCGGGCUUUAGCUUUUGGCUGGCACUCCGCA .....((((((.........(((((.((((((((..(((...((((((((((((..........)))..)))))))))))).)))))))).)))))........)))))).......... (-30.35 = -33.23 + 2.88)



| Location | 22,320,152 – 22,320,250 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -25.86 |
| Energy contribution | -27.18 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22320152 98 + 23771897 ---------------------AAGUGGCUGCCCAG-AAACUGGGUUCUAUUUGGGACCCAGAGUGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGACGGCACAGCACAUGU ---------------------..(((((((.((..-...(((((((((....))))))))).............(((((...(((....)))...)))))......))..)))).))).. ( -38.00) >DroSec_CAF1 78851 119 + 1 GAUAUGCAGCAGGAUGCAAAGAAGCGGCUGCCCAG-AAACUGGGUUCUAUUUGGGACCCAGAGUGUAGAGAAUCCGGCGAUGCUGGAGACGGAACCGCCGAAAGAUGCCACAGCACAUGU .....(((((....(((......))))))))....-...(((((((((....))))))))).((((.(.(.((((((((...(((....)))...)))))...)))..).).)))).... ( -43.00) >DroEre_CAF1 77219 120 + 1 GAUAUGCAGCAGGAUGCAAGGAAGCGGCUGCCCAGAGAAGUGGCUUCUAUUUGGGACCUAGAGGGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGAUGCCACAGCACAUGU ....(((.((((..(((......))).))))........(((((.(((((((...(((.....)))...)))).(((((...(((....)))...)))))..))).))))).)))..... ( -40.80) >DroYak_CAF1 57240 118 + 1 GAUAUGCAGUAGGAUGCAAGGAAGCGGCUGUUUAG-GAACUGGGUUCUAUUUGGUAC-GAGUGGGUAGAGAAUCUGGGGAUACUGGAGACGAAACUGCCAAAAGAUGCCACAGCAAAUGU ....((((......))))........(((((...(-((((...)))))....(((((-.....(((((.(..((..(.....)..))..)....)))))....).)))))))))...... ( -28.50) >consensus GAUAUGCAGCAGGAUGCAAGGAAGCGGCUGCCCAG_AAACUGGGUUCUAUUUGGGACCCAGAGGGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGAUGCCACAGCACAUGU ....((((......))))........((((.........(((((((((....))))))))).............(((((...(((....)))...)))))..........))))...... (-25.86 = -27.18 + 1.31)

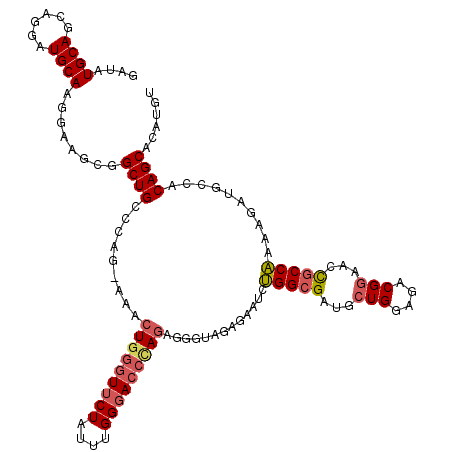
| Location | 22,320,170 – 22,320,290 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -41.94 |
| Consensus MFE | -34.26 |
| Energy contribution | -37.33 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22320170 120 + 23771897 UGGGUUCUAUUUGGGACCCAGAGUGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGACGGCACAGCACAUGUGUCCCUGCAUUGGCAUUUUCUUUUGCUUUCUUUUCUCUCU ((((((((....))))))))(((.(.((((((...((((...(((....)))...))))((((((.((((((.....))))))..(((....)))...))))))...)))))).).))). ( -44.10) >DroSec_CAF1 78890 120 + 1 UGGGUUCUAUUUGGGACCCAGAGUGUAGAGAAUCCGGCGAUGCUGGAGACGGAACCGCCGAAAGAUGCCACAGCACAUGUGUCCCUGCUCUGGCAUUAUCUUUUGCUUUCUUUUCCCUCU ((((((((....))))))))(((.(.((((((..(((((...(((....)))...)))))...(((((((.((((..........)))).)))))))..........)))))).).))). ( -47.00) >DroEre_CAF1 77259 119 + 1 UGGCUUCUAUUUGGGACCUAGAGGGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGAUGCCACAGCACAUGUGUCCCUGCUCCGGCAUUUUCUUUUGCUUUCUUUC-CCUCU .((.((((....)))))).((((((.((((((..(((((...(((....)))...))))).((((((((..((((..........))))..))))))))........)))))))-))))) ( -42.70) >DroYak_CAF1 57279 119 + 1 UGGGUUCUAUUUGGUAC-GAGUGGGUAGAGAAUCUGGGGAUACUGGAGACGAAACUGCCAAAAGAUGCCACAGCAAAUGUGUCCCUGCUUUGGCAUUUUCUUUUGCUUUCUUUUCCCUCU .(((........((((.-(((..(((((.(..((..(.....)..))..)....)))))..(((((((((.((((..........)))).)))))))))))).)))).......)))... ( -33.96) >consensus UGGGUUCUAUUUGGGACCCAGAGGGUAGAGAAUCUGGCGAUGCUGGAGACGGAACCGCCAAAAGAUGCCACAGCACAUGUGUCCCUGCUCUGGCAUUUUCUUUUGCUUUCUUUUCCCUCU ((((((((....))))))))(((((.((((((..(((((...(((....)))...))))).(((((((((.((((..........)))).)))))))))........)))))).))))). (-34.26 = -37.33 + 3.06)



| Location | 22,320,210 – 22,320,328 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22320210 118 - 23771897 AGAGAGGUUGAGCAAAAAGUAACAAAUAGUGUUGGGGC--AGAGAGAAAAGAAAGCAAAAGAAAAUGCCAAUGCAGGGACACAUGUGCUGUGCCGUCUUUUGGCGGUUCCGUCUCCAGCA ......((((.(((.....(((((.....))))).(((--(........................))))..))).((((((((.....)))((((((....))))))...))))))))). ( -27.16) >DroSec_CAF1 78930 118 - 1 AGAGAGGUUGAGCAAAAAGUAUCAAAUAGUGUUGGGGC--AGAGGGAAAAGAAAGCAAAAGAUAAUGCCAGAGCAGGGACACAUGUGCUGUGGCAUCUUUCGGCGGUUCCGUCUCCAGCA .......((((((.....)).))))....((((((...--(((.((((......((.(((((...(((((.((((.(......).)))).))))))))))..))..)))).))))))))) ( -34.90) >DroEre_CAF1 77299 119 - 1 AGAGAGGUUGAGCAAAAAGUAACAAAUAGUGUUGGGGAAAAGAGG-GAAAGAAAGCAAAAGAAAAUGCCGGAGCAGGGACACAUGUGCUGUGGCAUCUUUUGGCGGUUCCGUCUCCAGCA .............................(((((((((.....((-((..(....(((((((...(((((.((((.(......).)))).)))))))))))).)..)))).))))))))) ( -37.10) >DroYak_CAF1 57318 118 - 1 AGUAAGGUUGAGCAAAAUGUAGCAAAUAGUGUUGGGAC--AGAGGGAAAAGAAAGCAAAAGAAAAUGCCAAAGCAGGGACACAUUUGCUGUGGCAUCUUUUGGCAGUUUCGUCUCCAGUA .((.(.(((......))).).))........((((...--(((.((((..(....(((((((...(((((.((((((......)))))).)))))))))))).)..)))).))))))).. ( -28.70) >consensus AGAGAGGUUGAGCAAAAAGUAACAAAUAGUGUUGGGGC__AGAGGGAAAAGAAAGCAAAAGAAAAUGCCAAAGCAGGGACACAUGUGCUGUGGCAUCUUUUGGCGGUUCCGUCUCCAGCA .............................((((((.....(((.((((..(....(((((((...(((((.((((.(......).)))).)))))))))))).)..)))).))))))))) (-25.52 = -25.90 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:29 2006