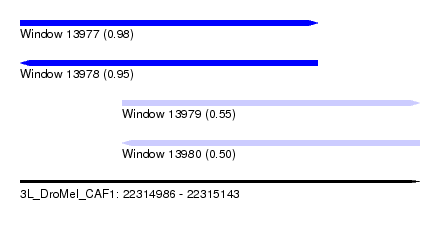
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,314,986 – 22,315,143 |
| Length | 157 |
| Max. P | 0.981716 |

| Location | 22,314,986 – 22,315,103 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -33.79 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314986 117 + 23771897 AUCCAUUACUGUUACAUCGCUGGAACCCAGCGAAGACGAAGUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAUUGAAAUGGAAUGGGUAGA---GAAUGGGUUU (((((((..((((...(((((((...))))))).)))).......(((..............((((((((.....))))))))(((((((.......))))))).))---)))))))).. ( -40.20) >DroSec_CAF1 73672 120 + 1 AUCCAUUACUGUUUCAUCGCUGGAACCCAGCGAAGGCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAACGGAAUGGGUAGAGGAGAAUGGGUUU (((((((.(((((((((((((((...))))))).(((((.......))..............((((((((.....)))))))))))...)))))))))))))))................ ( -42.90) >DroEre_CAF1 72025 117 + 1 AUCCAUUACUGUUUCAUCGCUGGAACCCAGCGAAGGCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGA---GAAUGGGUUU (((((((.(((((((((((((((...))))))).(((((.......))..............((((((((.....)))))))))))...)))))))))))))))...---.......... ( -40.40) >DroYak_CAF1 52139 117 + 1 AUCCAUUACUGUUUCAUCGCUUGAAUCCAGCGAAGGUGAAAUUGCCUCAACAACUGUCAUUUUGACAGCUGGCUCAGCUGUCAGCCCAAUAAAAGGGACUGGGUAGA---GAAUUGGUUU ..(((......((((((((((.((..(((((((.(((......)))))......((((.....))))))))).)))))...((((((.......))).)))))).))---))..)))... ( -32.20) >consensus AUCCAUUACUGUUUCAUCGCUGGAACCCAGCGAAGGCGAAAUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGA___GAAUGGGUUU (((((((.(((((((((((((((...))))))).((((((((.((..........)).)))))(((((((.....))))))).)))...)))))))))))))))................ (-33.79 = -34.60 + 0.81)


| Location | 22,314,986 – 22,315,103 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -31.64 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314986 117 - 23771897 AAACCCAUUC---UCUACCCAUUCCAUUUCAAUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAACUUCGUCUUCGCUGGGUUCCAGCGAUGUAACAGUAAUGGAU ....(((((.---((..((((((.......))))))..))..(((((((.(((((((.....))))))))))((((...))))....(((((((...))))))).....))))))))).. ( -36.40) >DroSec_CAF1 73672 120 - 1 AAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGCCUUCGCUGGGUUCCAGCGAUGAAACAGUAAUGGAU ..................((((((.((((((((..((((..((((.(((((((((((.....)))))))...(((((......))))))))).)))).)))))))))))).).))))).. ( -41.90) >DroEre_CAF1 72025 117 - 1 AAACCCAUUC---UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGCCUUCGCUGGGUUCCAGCGAUGAAACAGUAAUGGAU ..........---.....((((((..(((((((..((((..((((.(((((((((((.....)))))))...(((((......))))))))).)))).)))))))))))..).))))).. ( -38.40) >DroYak_CAF1 52139 117 - 1 AAACCAAUUC---UCUACCCAGUCCCUUUUAUUGGGCUGACAGCUGAGCCAGCUGUCAAAAUGACAGUUGUUGAGGCAAUUUCACCUUCGCUGGAUUCAAGCGAUGAAACAGUAAUGGAU ...(((....---......((((((........))))))...((((.((((((((((.....))))))).....)))..(((((...(((((.......))))))))))))))..))).. ( -35.70) >consensus AAACCCAUUC___UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAUUUCGCCUUCGCUGGGUUCCAGCGAUGAAACAGUAAUGGAU ....(((((....((..((((((.......))))))..))..(((((((.(((((((.....)))))))))).(((........)))(((((((...))))))).....))))))))).. (-31.64 = -32.33 + 0.69)



| Location | 22,315,026 – 22,315,143 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -23.63 |
| Energy contribution | -24.19 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22315026 117 + 23771897 GUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAUUGAAAUGGAAUGGGUAGA---GAAUGGGUUUGAGGAUGAGGAGCGAAAGAAACCGGCAACACAAAGAGCGA (((.((((((..(((..(((((((((((((.....))))))).(((((((.......)))))))..)---)))))))))))))).((.((..(....)...))(....).))...))).. ( -36.00) >DroSec_CAF1 73712 120 + 1 AUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAACGGAAUGGGUAGAGGAGAAUGGGUUUGAGGAUGAGGAGCGAAAGAAACCGGCAACACAAAGAGCGA ....((((((..(((..(((((((((((((.....))))))).(((((...........))))).....)))))))))))))))....((..(....)...))(....)........... ( -32.40) >DroEre_CAF1 72065 116 + 1 AUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGA---GAAUGGGUUUGGG-AUGAGGAGUAAAGGGAACCGGCAAAACAAAGAGGGA ....((((........((((((((((((((.....))))))).(((((...........)))))..)---)))))).((((..-.((.((...........))..))...)))))))).. ( -29.10) >DroYak_CAF1 52179 117 + 1 AUUGCCUCAACAACUGUCAUUUUGACAGCUGGCUCAGCUGUCAGCCCAAUAAAAGGGACUGGGUAGA---GAAUUGGUUUGAGAACGAGCAGUACAAGAAACCGGCAAAACAAAGAAGGA .(((((....(((.(.((....(((((((((...)))))))))(((((...........))))).))---.).)))(((((....))))).............)))))............ ( -32.30) >consensus AUUACCUCAACAACUGUCAUUUUGACAGCUCGCUCAGCUGUCAGCCCAAUGAAAUGGAAUGGGUAGA___GAAUGGGUUUGAGGAUGAGGAGCAAAAGAAACCGGCAAAACAAAGAGCGA ....((((......((((....((((((((.....))))))))(((((...........)))))...........(((((..................))))))))).......)))).. (-23.63 = -24.19 + 0.56)


| Location | 22,315,026 – 22,315,143 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -23.08 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22315026 117 - 23771897 UCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUC---UCUACCCAUUCCAUUUCAAUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAC ...........((((((........((((((.......(((..((((((.---.................)))))).))).....))))))((((((.....))))))......)))))) ( -28.47) >DroSec_CAF1 73712 120 - 1 UCGCUCUUUGUGUUGCCGGUUUCUUUCGCUCCUCAUCCUCAAACCCAUUCUCCUCUACCCAUUCCGUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAU .(((.....)))(((((........((((((.......(((..((((.........................)))).))).....))))))((((((.....))))))......))))). ( -26.21) >DroEre_CAF1 72065 116 - 1 UCCCUCUUUGUUUUGCCGGUUCCCUUUACUCCUCAU-CCCAAACCCAUUC---UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAU ..((((...((((((.((((((.(((..((..(((.-(((((........---..................))))).))).))..))).)))))).))))))(((....))))))).... ( -26.37) >DroYak_CAF1 52179 117 - 1 UCCUUCUUUGUUUUGCCGGUUUCUUGUACUGCUCGUUCUCAAACCAAUUC---UCUACCCAGUCCCUUUUAUUGGGCUGACAGCUGAGCCAGCUGUCAAAAUGACAGUUGUUGAGGCAAU ............(((((.............(((((((.(((..(((((..---.................)))))..))).))).))))((((((((.....))))))))....))))). ( -32.61) >consensus UCCCUCUUUGUGUUGCCGGUUUCUUUCACUCCUCAUCCUCAAACCCAUUC___UCUACCCAUUCCAUUUCAUUGGGCUGACAGCUGAGCGAGCUGUCAAAAUGACAGUUGUUGAGGUAAU ............((((((((((..................)))))............((((((.......)))))).((((((((.....))))))))................))))). (-23.08 = -23.52 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:16 2006