
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,314,125 – 22,314,405 |
| Length | 280 |
| Max. P | 0.905468 |

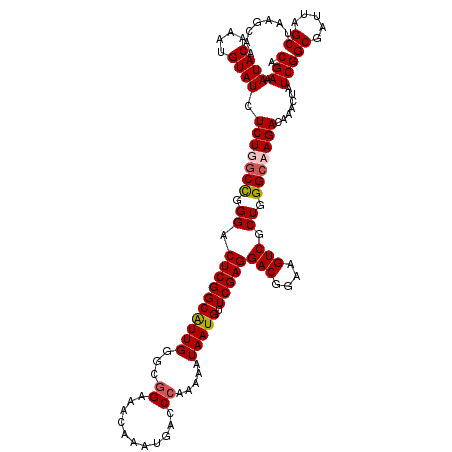
| Location | 22,314,125 – 22,314,245 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314125 120 - 23771897 ACGAAAUACAAAUGUAUCUCUUGCCGGGACUCGGCGUUGGGCGGAAACAAAUGACCCAAAAAUAAUGUUCGAGGACGGAAGUCGCUGGGCAAGACAAACUAUCGGCGAUUAGCUAAGCAA .(((.((((....)))).(((((((.((.((((((((((((((........)).))))......))).)))))(((....))).)).))))))).......)))((.....))....... ( -35.80) >DroSec_CAF1 72811 116 - 1 ACGAAAUACAAAUGUAUCUCUUGCCGGGACUCGGCGUUG----GAAACAAAUGACCCAAAAAUAAUGUUCGAGGACGGAAGUCGCUGGGCAAGACAAACUAUCGGCGAUUAGCUAAGCAA .(((.((((....)))).((((((((((..(((...(((----....))).))))))..............(((((....))).)).))))))).......)))((.....))....... ( -32.30) >DroEre_CAF1 71169 120 - 1 ACGAAAUACAAAUGUAUCUCUGGCCGGGACUCGGCAUUGGGCGGAAACAAAUGACCCAAAAAUAAUGUUCGAGGACGGAAGUCGCUGGGCAAGACAAACUAUCGGCGAUUAGCUAAGCAA .(((.((((....)))).(((.(((.((.((((((((((((((........)).))))......))).)))))(((....))).)).))).))).......)))((.....))....... ( -32.10) >DroYak_CAF1 51308 120 - 1 ACGAAAUACAAAUGUAUCUCUGUCUGGGACUCGGCAUUGAGUGGAAACAAAUGACCCAAAAAUAAUGUUCGAGGACGGAAGUCUCUGGGCAAGACAAACUAUCGGCGAUUAGCUAAGGAA ..((.((((....)))).))(((((((((((((....)))))(....)......)))........((((((..(((....)))..))))))))))).......(((.....)))...... ( -34.20) >consensus ACGAAAUACAAAUGUAUCUCUGGCCGGGACUCGGCAUUGGGCGGAAACAAAUGACCCAAAAAUAAUGUUCGAGGACGGAAGUCGCUGGGCAAGACAAACUAUCGGCGAUUAGCUAAGCAA .(((.((((....)))).(((((((.((.((((((((((...((...........)).....))))).)))))(((....))).)).))))))).......)))((.....))....... (-28.66 = -29.23 + 0.56)

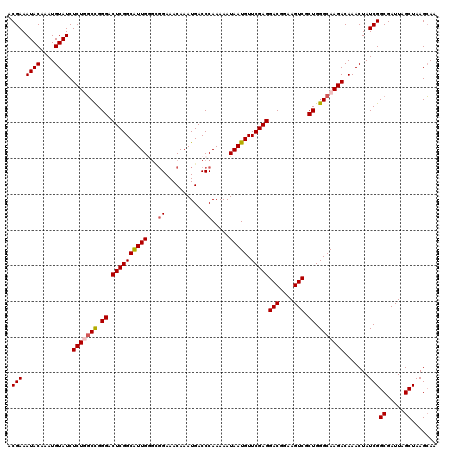
| Location | 22,314,165 – 22,314,285 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314165 120 + 23771897 UUCCGUCCUCGAACAUUAUUUUUGGGUCAUUUGUUUCCGCCCAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGAUGUGGCUCGGCGA .....................((((((...........))))))(((((((((.(((((.(((((((....))))))).))))..(((........))).........).))))))))). ( -38.80) >DroSec_CAF1 72851 116 + 1 UUCCGUCCUCGAACAUUAUUUUUGGGUCAUUUGUUUC----CAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGA ....(.(((.(((......))).))).).........----...(((((((((.((((..(((((((....)))))))...(((.(((........)))..))).)))).))))))))). ( -38.60) >DroEre_CAF1 71209 120 + 1 UUCCGUCCUCGAACAUUAUUUUUGGGUCAUUUGUUUCCGCCCAAUGCCGAGUCCCGGCCAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGCUGCAAAUAGAGCUGUGGCUCGCCGA ........(((..........((((((...........))))))...((((((.(((((.(((((((....))))))).(((..............)))....).)))).)))))).))) ( -30.04) >DroYak_CAF1 51348 120 + 1 UUCCGUCCUCGAACAUUAUUUUUGGGUCAUUUGUUUCCACUCAAUGCCGAGUCCCAGACAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGA ....(.(((.(((......))).))).)................(((((((((.(((.(.(((((((....)))))))...(((.(((........)))..))).)))).))))))))). ( -33.50) >consensus UUCCGUCCUCGAACAUUAUUUUUGGGUCAUUUGUUUCCGCCCAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGA ....(.(((.(((......))).))).)................(((((((((.((((..(((((((....)))))))...(((.(((........)))..))).)))).))))))))). (-31.06 = -31.62 + 0.56)


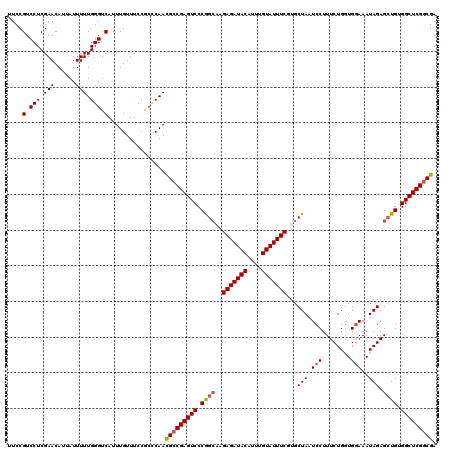
| Location | 22,314,205 – 22,314,325 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -44.64 |
| Consensus MFE | -37.12 |
| Energy contribution | -38.25 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314205 120 + 23771897 CCAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGAUGUGGCUCGGCGAGCGGUCUCAUCCAUCAGGGCAACCUCAACGGGGGGAUUAG ...((((((.....))))..(((((((....))))))))).((((((((..((((((((....(((((...((((...)))).))))).)))))))((....)).....)..)))))))) ( -48.20) >DroSec_CAF1 72888 119 + 1 -CAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGAGCGGUCUCAUCCAUCAGGGCAACCUCAACGGGGGGAUUAG -..((((((.....))))..(((((((....))))))))).((((((((..((((((((....((((....))))(((.....)))...)))))))((....)).....)..)))))))) ( -48.60) >DroEre_CAF1 71249 120 + 1 CCAAUGCCGAGUCCCGGCCAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGCUGCAAAUAGAGCUGUGGCUCGCCGAGCGGUCUCAUCCAUCAGCGCAACCUCAACGGGCGGAUUAU ......(((...((((....(((((((....)))))))(((((..........(((((.....((((....)))).....)))))..........)))))........)))))))..... ( -35.85) >DroYak_CAF1 51388 119 + 1 UCAAUGCCGAGUCCCAGACAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGAGCGGUCCCAUCCAUUAGGGCAACC-CAACGGGGGGAUUAG ....(((((((((.(((.(.(((((((....)))))))...(((.(((........)))..))).)))).)))))))))...((((((.(((.((.((....))-.)).))))))))).. ( -45.90) >consensus CCAACGCCGAGUCCCGGCAAGAGAUACAUUUGUAUUUCGUGCUAAUCCUUUCUGGUGGAAAUAGAGCUGUGGCUCGGCGAGCGGUCUCAUCCAUCAGGGCAACCUCAACGGGGGGAUUAG ....(((((((((.((((..(((((((....)))))))...(((.(((........)))..))).)))).)))))))))...((((((.(((...(((....)))....))))))))).. (-37.12 = -38.25 + 1.12)

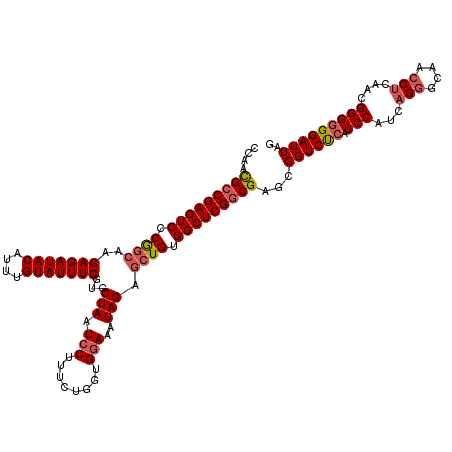
| Location | 22,314,285 – 22,314,405 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -31.14 |
| Energy contribution | -32.95 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22314285 120 - 23771897 AUAAUCAUUUUCAAAUGCAUCCUUAGAUGGCACAAACUUCUACCGUUGUCAGUCAGGGAAGAAGUUUAUCGGGGGCCAAACUAAUCCCCCCGUUGAGGUUGCCCUGAUGGAUGAGACCGC .................(((((...((..((.............))..)).(((((((...((.((((.((((((...........)))))).)))).)).))))))))))))....... ( -36.52) >DroSec_CAF1 72967 120 - 1 AUAAUCAUUUUCAAAUGCAUCCUUAGACGGCACAAACUUCUGCCGUUGUCAGUCAGGGAAGCAGUUUAUCGGGGGCCAAACUAAUCCCCCCGUUGAGGUUGCCCUGAUGGAUGAGACCGC .................(((((...(((((((........)))))))....(((((((.(((...(((.((((((...........)))))).))).))).))))))))))))....... ( -42.40) >DroEre_CAF1 71329 120 - 1 AUAAUCAUUUUCAAAUGCAUCCCCAGACGGCACAAACCUCUGCCGCUGUCAGUCAGGGAAGCAGUUUAUCGGGGGCUAAAAUAAUCCGCCCGUUGAGGUUGCGCUGAUGGAUGAGACCGC ........((((...(((.(((((((.(((((........)))))))).......)))).)))(((((((((((((...........))))((.......)).))))))))))))).... ( -35.11) >DroYak_CAF1 51468 119 - 1 AUAAUGAUUUUCAAAUGCAUCCUCAGACGGCACAAAUUUCUGCCGUUGUCAGUCAGGGCAGCAGUUUAUAGGGGGCUAAACUAAUCCCCCCGUUG-GGUUGCCCUAAUGGAUGGGACCGC ...............(.(((((...(((((((........))))))).......((((((((.....(..(((((..........)))))..)..-.))))))))...))))).)..... ( -40.20) >consensus AUAAUCAUUUUCAAAUGCAUCCUCAGACGGCACAAACUUCUGCCGUUGUCAGUCAGGGAAGCAGUUUAUCGGGGGCCAAACUAAUCCCCCCGUUGAGGUUGCCCUGAUGGAUGAGACCGC .................(((((...(((((((........)))))))....(((((((.(((...(((.((((((...........)))))).))).))).))))))))))))....... (-31.14 = -32.95 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:13 2006