
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,312,707 – 22,312,907 |
| Length | 200 |
| Max. P | 0.930187 |

| Location | 22,312,707 – 22,312,827 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.16 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22312707 120 - 23771897 AUUGGCUGCCUAAUGAAUUAUGCAUGGGCAGUCCAAAAAGCACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUAUAAUGCAUAUUAAGUGCGAACAACGAAUCGUAUUGAACGAGG .(((((((((((.((.......))))))))).))))...(((((((..............)))...........(((((.....)))))..))))..........((((.....)))).. ( -27.64) >DroSec_CAF1 71422 119 - 1 AUUGGCUGCCUAAUGUAAUAUGCAUGGGCAGCCCAAAAAG-ACGACACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACAAUGCAUAUUAAGUGCGAACAACGAAUCGUAUCGAACGAGG ...(((((((((.(((.....)))))))))))).......-........((((............................(((((.....))))).....(((......)))...)))) ( -27.90) >DroSim_CAF1 53135 119 - 1 AUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG-ACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACAAUGCAUAUUAAGUGCGAACAACGAAUCGUAUCGAACGAGG ...(((((((((.((.......)))))))))))...((((-..((....))..))))....((..................(((((.....))))).........((((.....)))))) ( -26.80) >DroEre_CAF1 69781 119 - 1 AUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAGAAG-GCGGCACGCCUCCUUUACAGCUAGAUAAAUCAAAAUACAAUGCAUAUUAAGUGCGAGCGACGAAUCGUAUCGAACGAGG ...(((((((((.((.......)))))))))))...((((-(.((....)).)))))...(((.((....)).........(((((.....))))))))......((((.....)))).. ( -33.20) >DroYak_CAF1 49898 119 - 1 AUUGGCUGCCUAAUGAAAUAUGCAAAGGCAGUCCAAAAAG-GCGGCACGCCUCCUUUACAGCUAGAUAAAUCAAAAUACAAUGCAUAUUAAGUGCGAACAACGAAUCGUAUCGAACGAGG .((((((((((..............)))))).))))((((-(.((....)).)))))........................(((((.....))))).........((((.....)))).. ( -27.24) >consensus AUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG_ACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACAAUGCAUAUUAAGUGCGAACAACGAAUCGUAUCGAACGAGG ...(((((((((.((.......)))))))))))................((((............................(((((.....))))).....(((......)))...)))) (-24.52 = -24.16 + -0.36)

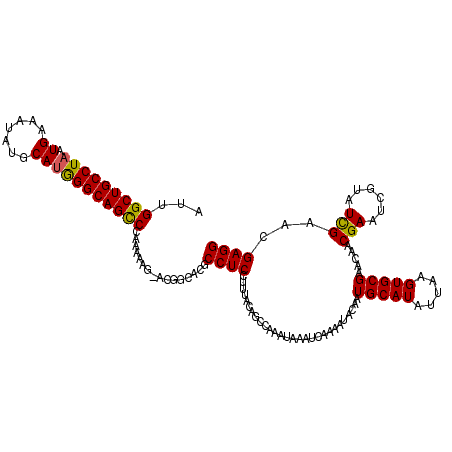
| Location | 22,312,747 – 22,312,867 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22312747 120 - 23771897 UUGGCAGACUGUUCAAGACGCAGGGCCGGGGGAGGAAUUCAUUGGCUGCCUAAUGAAUUAUGCAUGGGCAGUCCAAAAAGCACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUAUA (((((...((((.......))))....((((((((......(((((((((((.((.......))))))))).))))...((...))...))))))))...)))))............... ( -36.30) >DroSec_CAF1 71462 119 - 1 CUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGUAAUAUGCAUGGGCAGCCCAAAAAG-ACGACACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACA .((((...((((.......))))......((((((...((...(((((((((.(((.....))))))))))))(.....)-..))....)))))).....))))................ ( -35.40) >DroSim_CAF1 53175 119 - 1 CUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG-ACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACA .((((...((((.......))))(((.((((((....)))...(((((((((.((.......))))))))))).......-.)))...))).........))))................ ( -33.60) >DroEre_CAF1 69821 119 - 1 UUGGCAGACAAUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAGAAG-GCGGCACGCCUCCUUUACAGCUAGAUAAAUCAAAAUACA (((((.................(((..((....))..)))...(((((((((.((.......)))))))))))...((((-(.((....)).)))))...)))))............... ( -32.50) >DroYak_CAF1 49938 119 - 1 UUGGCAGACUAUUCAAGACACAGUGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAAAGGCAGUCCAAAAAG-GCGGCACGCCUCCUUUACAGCUAGAUAAAUCAAAAUACA ..(((.((....))........(((((((.(((....))).((((((((((..............)))))).))))...)-).))))))))............................. ( -30.34) >consensus UUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG_ACGGCACGCCUCCUUUACAGCCAAAUAAAUCAAAAUACA .((((...((((.......))))......((((((........(((((((((.((.......))))))))))).........((...)))))))).....))))................ (-28.22 = -28.22 + 0.00)



| Location | 22,312,787 – 22,312,907 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -37.79 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.698047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22312787 120 - 23771897 GACCCAAGACUUGUACCACUUUAUCGAAUCGAUAAGCCCCUUGGCAGACUGUUCAAGACGCAGGGCCGGGGGAGGAAUUCAUUGGCUGCCUAAUGAAUUAUGCAUGGGCAGUCCAAAAAG ...............((..(((((((...))))))).((((((((...((((.......)))).)))))))).))......(((((((((((.((.......))))))))).)))).... ( -39.70) >DroSec_CAF1 71501 120 - 1 GGCCCCAGACUUGUACUACUUUAUCGAAUCGAUAAGCCCCCUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGUAAUAUGCAUGGGCAGCCCAAAAAG (((((..(.((((.((....((((((...))))))(((....))).....)).)))).)...)))))((....))........(((((((((.(((.....))))))))))))....... ( -42.70) >DroSim_CAF1 53214 120 - 1 GGCCCCAGACUUGUACUACUUUAUCGAAUCGAUAAGCCCCCUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG (((((..(.((((.((....((((((...))))))(((....))).....)).)))).)...)))))((....))........(((((((((.((.......)))))))))))....... ( -40.70) >DroEre_CAF1 69860 119 - 1 GCCCG-AGACUUGUACUACCAUAUCGAUUCGAUAAGCCCCUUGGCAGACAAUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAGAAG ((((.-.(.((((........(((((...))))).(((....)))........)))).)...)))).((....))..(((...(((((((((.((.......)))))))))))...))). ( -37.20) >DroYak_CAF1 49977 120 - 1 GGCCGAAGACUUGUACUACUAUAUCGAAUCGAUAAGCCCCUUGGCAGACUAUUCAAGACACAGUGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAAAGGCAGUCCAAAAAG ((((.....((((........(((((...))))).(((....)))........)))).....).)))((....))......((((((((((..............)))))).)))).... ( -28.64) >consensus GGCCCAAGACUUGUACUACUUUAUCGAAUCGAUAAGCCCCUUGGCAGACUGUUCAAGACGCAGGGCCCCGGGAGGAAUUCAUUGGCUGCCUAAUGAAAUAUGCAUGGGCAGCCCAAAAAG ..((...............(((((((...))))))).(((..(((...((((.......)))).)))..))).))........(((((((((.((.......)))))))))))....... (-30.32 = -30.80 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:05 2006