
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,311,332 – 22,311,645 |
| Length | 313 |
| Max. P | 0.889796 |

| Location | 22,311,332 – 22,311,449 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22311332 117 + 23771897 AAAAGUGGUCAACAUGACAUGGUUGACAUGCGACAUUAGCAAAGGCA--CAACACAAGGCUUCCUUGGGA-CCCCUAACCCUUCCCUCUUUGUGCGCCAACCCAAGAACCCAGCUACCCA ...(((.((((((........)))))).(((.......)))..(((.--...(((((((.......(((.-.......)))......))))))).)))..............)))..... ( -25.12) >DroSec_CAF1 70037 119 + 1 AAAAGUGGUCAACAUGACAACGUUGACAUGCGACAUUAGCAGAGGCACACAACACAAGGCUUCCUUGGGA-CCCCUAACCCUUCCCUCUCUGUGUGCCAACCCAAGAACCCAGCUAACCG ....(((((((((........)))))).)))....(((((...((((((((....((((...))))((((-...........))))....)))))))).......(....).)))))... ( -31.20) >DroEre_CAF1 68460 94 + 1 AAAAGUGGUCAACAUGACAACGUUGACAUGCGACAUUAGCAGAGGCA--CAACACAGGGGUUCCUUGAGGUCCCCCAACCCU------------------------AGCCCAGCUACCCA ....(((((((((........)))))).))).....((((.(.(((.--.......((((..((....))..))))......------------------------.)))).)))).... ( -26.76) >DroYak_CAF1 48526 117 + 1 AAAAGUGGUCAACAUGACAACGUUGACAUGCGACAUUAGCAGAGGUA--CAACACAGGGGUUCCUUGGGAUCCCCUUACACUUCCCUCUCUGUGU-CCAACACAAGAACCCAGCUACCCA ....(((((((((........)))))).((.(((((.((.(((((..--......(((((.(((...))).)))))........)))))))))))-))).)))................. ( -36.19) >consensus AAAAGUGGUCAACAUGACAACGUUGACAUGCGACAUUAGCAGAGGCA__CAACACAAGGCUUCCUUGGGA_CCCCUAACCCUUCCCUCUCUGUGU_CCAACCCAAGAACCCAGCUACCCA ....(((((((((........)))))).(((.......)))...........)))..((((((.(((((....))))).............((......))....))))))......... (-18.85 = -19.85 + 1.00)



| Location | 22,311,449 – 22,311,568 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -35.79 |
| Energy contribution | -35.72 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22311449 119 - 23771897 CUUU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGUGGGUAUUUUUCUUAGGCGAGGAGCACGCAAGGAGCUCAGAAAGGAGGUGGC ((((-(((..(((((((((((((((((((((.((((.(((.........))).)))).)))))))).)))))))).........)))))((((.(....)..)))))))))))....... ( -44.80) >DroSec_CAF1 70156 120 - 1 CUUUUUCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGUGGGCAUUUUUCUUGGGCGAAGAGCACGCAAGGAGCUCGGAAAGAAGAUGGC .(((((((((((((.((((((((((((((((.((((.(((.........))).)))).)))))))).))))))))......))))))).((((.(....)..)))).))))))....... ( -46.40) >DroEre_CAF1 68554 117 - 1 CUUU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGGGCAUUUUUCAUAGGCGAGGAGCACGCAAGGAGCUCGGAAAGGAA--GGC ((((-(((.((((((((((((.(((...((.(((((..((..((((((((((....)))))...)).)))..))..))))))).)))..))))))))...))))..)))))))..--... ( -45.00) >DroYak_CAF1 48643 116 - 1 CUGU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUAAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGCGCAUUUUUCAUAGGCGAGGAGCACGCAAUGAGCUCGGAAAGGAA---CC ((.(-(((.((((((((((((.(((...((.(((((....(((((((.((((....))))((.....)))))))))))))))).)))..))))))))...))))..)))).))..---.. ( -42.20) >consensus CUUU_UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGGGCAUUUUUCAUAGGCGAGGAGCACGCAAGGAGCUCGGAAAGGAA__GGC .....(((.((((((((((((.(((.......((((..((...(((((((((....))))).))))......))..))))....)))..))))))))...))))..)))........... (-35.79 = -35.72 + -0.06)


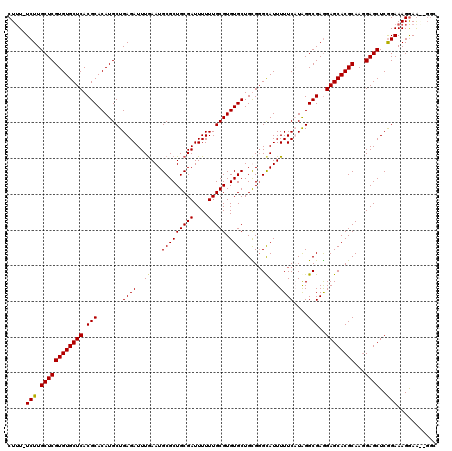
| Location | 22,311,489 – 22,311,608 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -34.59 |
| Energy contribution | -34.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22311489 119 - 23771897 GGAGCAUUUGAUAACAUGUUUUGCCAACCGGCCCGCACUGCUUU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGUGGGUAUUUUUC .(((((...((.(((((((...(((....)))..))).)).)).-)).)))))..((((((((((((((((.((((.(((.........))).)))).)))))))).))))))))..... ( -39.20) >DroSec_CAF1 70196 120 - 1 GGAGCAUUUGAUAACAUGUUUUGCCAACCGGCCCGCAUCGCUUUUUCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGUGGGCAUUUUUC .(((((...((....((((...(((....)))..)))).....))...)))))..((((((((((((((((.((((.(((.........))).)))).)))))))).))))))))..... ( -40.40) >DroEre_CAF1 68592 119 - 1 GGAGCAUUUGAUAACAUGUUUUGCCAGCCGGCCCGAACCGCUUU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGGGCAUUUUUC .(((((...((......((((.(((....)))..))))......-)).)))))..((((((.(((((((((.((((.(((.........))).)))).)))))))).).))))))..... ( -37.20) >DroYak_CAF1 48680 119 - 1 GGAGCAUUUGAUAACAUGUUUUGGCAACCAGCCCGCACCGCUGU-UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUAAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGCGCAUUUUUC .(((((...((((.(.(((...(((.....))).)))..).)))-)..)))))((((((.((.((((((((.((((((............))))))..)))))))))).))))))..... ( -36.70) >consensus GGAGCAUUUGAUAACAUGUUUUGCCAACCGGCCCGCACCGCUUU_UCUUGCUCGUGUGCUCACGCACAUGCUGAGAUUUGAAUGCGCUGCGAUUUUUUGCGUGUGCUGCGGGCAUUUUUC (((((((........)))))))........(((((((........((((...(((((((....)))))))..)))).......(((((((((....))))).)))))))))))....... (-34.59 = -34.65 + 0.06)


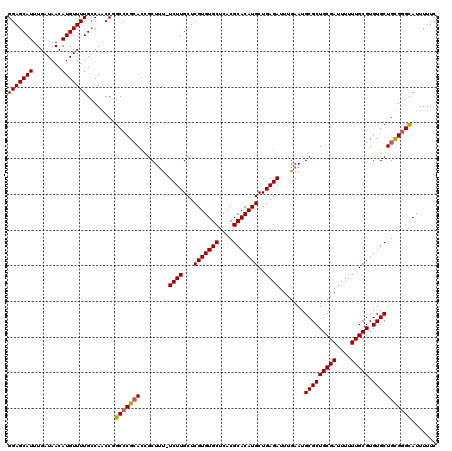
| Location | 22,311,529 – 22,311,645 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -27.66 |
| Energy contribution | -31.10 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22311529 116 + 23771897 CAAAUCUCAGCAUGUGCGUGAGCACACGAGCAAGA-AAAGCAGUGCGGGCCGGUUGGCAAAACAUGUUAUCAAAUGCUCCGACGCACCGCAAAUUGCCAGC-CGGACCACGACCACGA-- ....(((..((.(((((....)))))...)).)))-......((((((((((((((((((....((....))..(((......))).......))))))))-))).)).))..)))..-- ( -40.10) >DroSec_CAF1 70236 111 + 1 CAAAUCUCAGCAUGUGCGUGAGCACACGAGCAAGAAAAAGCGAUGCGGGCCGGUUGGCAAAACAUGUUAUCAAAUGCUCCGACGCACCGCAAAUUGCCAGC-CGGACCA------CGA-- .........((((((((....))))....((........)).))))((.(((((((((((....((....))..(((......))).......))))))))-))).)).------...-- ( -39.90) >DroEre_CAF1 68632 118 + 1 CAAAUCUCAGCAUGUGCGUGAGCACACGAGCAAGA-AAAGCGGUUCGGGCCGGCUGGCAAAACAUGUUAUCAAAUGCUCCGACGCACCGCAAAUUUGCAGC-CGGACCACGACCACGACC ....(((..((.(((((....)))))...)).)))-...(.(((.(((((((((((.((((...((....))..(((......))).......))))))))-))).)).))))).).... ( -39.30) >DroYak_CAF1 48720 117 + 1 UAAAUCUCAGCAUGUGCGUGAGCACACGAGCAAGA-ACAGCGGUGCGGGCUGGUUGCCAAAACAUGUUAUCAAAUGCUCCGACGCACCGCAAAUUGCCAGCCCGGACCACGACCACGA-- ....(((..((.(((((....)))))...)).)))-...(.(((.((((((((((((.......((....))..(((......)))..)))....))))))))).))).)........-- ( -40.60) >consensus CAAAUCUCAGCAUGUGCGUGAGCACACGAGCAAGA_AAAGCGGUGCGGGCCGGUUGGCAAAACAUGUUAUCAAAUGCUCCGACGCACCGCAAAUUGCCAGC_CGGACCACGACCACGA__ .........((((((((....))))....((........)).))))((.(((((((((((....((....))..(((......))).......)))))))).))).))............ (-27.66 = -31.10 + 3.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:18:02 2006